.png)
.png)
The complete guide to health data standardisation

Hannah Gaimster, PhD
In this article:
- What is health data standardisation?
- Why is health data standardisation important?
- What are the benefits of standardising health data?
- Enabling health data standardisation - technical challenges and solutions
- Key players driving health data standardisation
- Health data standardisation in action: enabling data linkage
- Health data standardisation: one part of the end-to-end data analysis pipeline
What is health data standardisation?
Let’s start with some background and context: healthcare is one of the most data-rich industries with exponential growth in data. This health data is needed to solve crucial scientific questions (eg, causes of rare diseases or the efficacy of a new drug). The data can come from a wide variety of sources. As a result, the data can exist in many different formats and be stored in many different places.
THE PROBLEM: Health data cannot be effectively combined for analysis
A significant barrier to harnessing the full potential of health data is that individual organisations adopt vastly different data standards to collect and store data. This has led to unstructured health datasets, creating huge reproducibility crises and impeding data interoperability.
THE SOLUTION: Health data standardisation
Essential health data often takes the form of large, unstructured datasets, but health data standardisation enables this data to be brought together and be interoperable.
Common Data Models (CDMs) are being utilised more widely in the healthcare sector to overcome the lack of consistency in health data

Health data standardisation refers to bringing data into an agreed-upon common format that allows for collaborative analysis.
The role of common data models in enabling data standardisation
Adopting CDMs, a standard collection of extensible schemas that provides common terminologies and formats, enables researchers to overcome the challenges of unstructured clinical datasets.
Collaborative health research on data across nations, sources, and systems is made possible by the standard approach to health data transformation provided by CDMs. Combining, accessing and analysing information is simpler when all health data are organised following a single worldwide standard.
Stakeholder-driven initiatives are progressing in implementing frameworks and building networks to support this mission. Examples of CDMs for clinical data are the:
What is OMOP?
OMOP is an open community data standard created to standardise observational data formats and content and to facilitate quick analyses. The OHDSI standardised vocabulary is a key part of the OMOP CDM. The OHDSI vocabularies enable standard analytics and allow the organisation and standardisation of medical terms to be used across the various clinical domains of the OMOP CDM.
What is CDISC?
OMOP is an open community data standard created to standardise observational data formats and content and to facilitate quick analyses. The OHDSI standardised vocabulary is a key part of the OMOP CDM. The OHDSI vocabularies enable standard analytics and allow the organisation and standardisation of medical terms to be used across the various clinical domains of the OMOP CDM.
What is FHIR?
OMOP is an open community data standard created to standardise observational data formats and content and to facilitate quick analyses. The OHDSI standardised vocabulary is a key part of the OMOP CDM. The OHDSI vocabularies enable standard analytics and allow the organisation and standardisation of medical terms to be used across the various clinical domains of the OMOP CDM.
Why is health data standardisation important?
It is increasingly important that health data undergo standardisation for various reasons.
The world's health data assets are currently not being utilised to their full potential
There are several reasons for this including:
- Diverse data sources
-
Health data comes from various sources, including electronic health record (EHR) systems and medical equipment. These sources may employ various models, schemas, and formats to collect clinical data. This makes it difficult to standardise and integrate the data.
- Data volume and complexity
-
Healthcare organisations frequently need to process enormous volumes of data (terabytes in volume), comprising both structured and unstructured data (eg, clinical notes) and requiring various standardisation approaches.
- Data heterogeneity
-
Health data is recorded in various forms, languages, and units. This is known as data heterogeneity. Standardising terminology, measurement units, and health codes is difficult when working with data from many organisations and places.
- Interoperability
-
Designing flexible application programming interfaces (APIs) that enable communication between many different datasets is technically challenging.
- Quality and consistency
-
When clinical data is standardised, it is crucial to guarantee data quality and consistency. Erroneous clinical or research judgements can be made due to inaccurate health data.
- Evolving data standards
-
To account for new knowledge, technological developments, and adjustments in healthcare practices, healthcare data standards and terminology are constantly changing.
- Security and privacy
-
Organisations must uphold patient privacy and follow data security laws (such as HIPAA) when standardising clinical data. Moving the data further increases the security risks by allowing third parties access
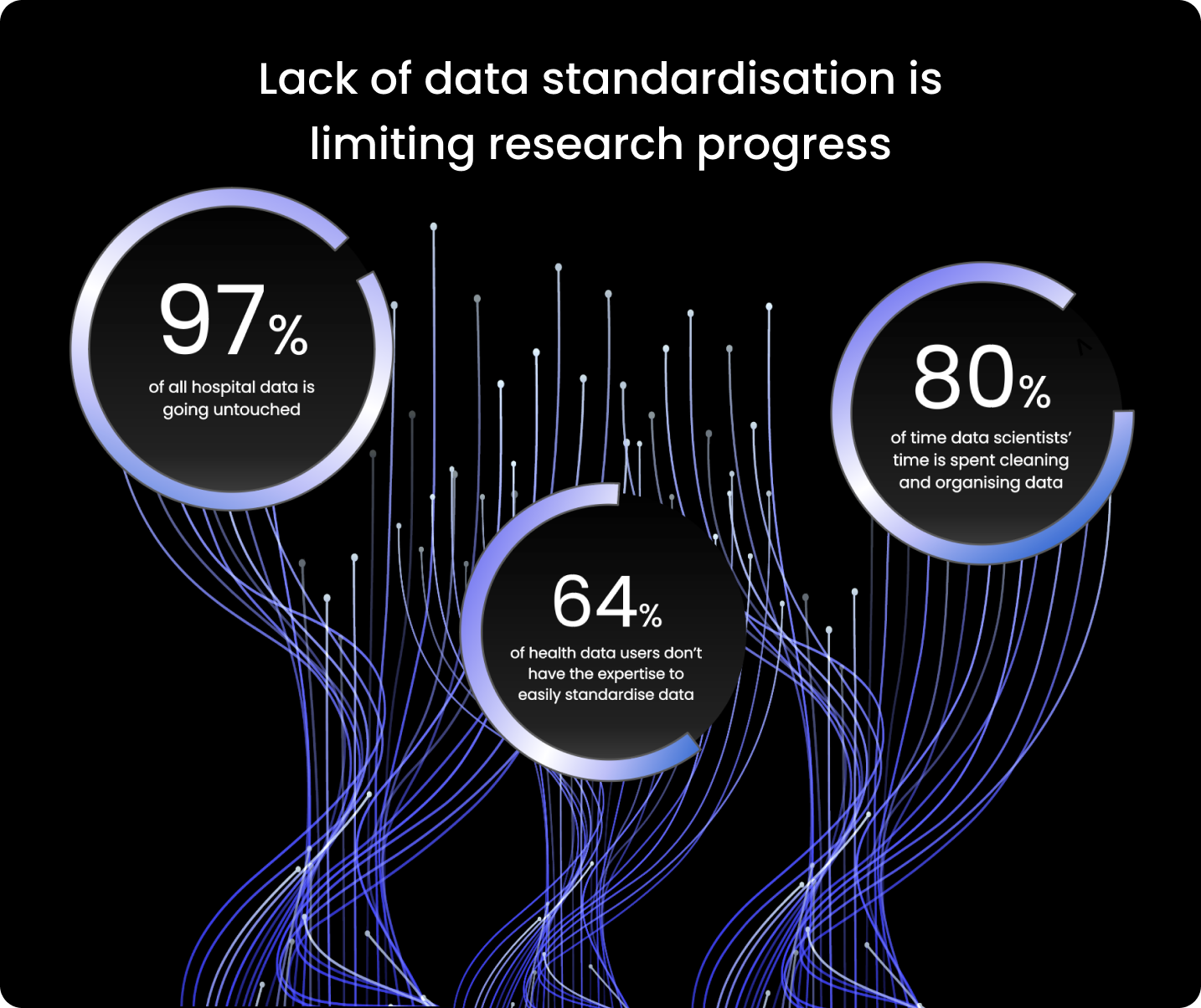
The World Economic Forum estimates that
97% of hospital data goes unused.
Since the majority of users of health data (64%) lack the knowledge necessary to standardise data quickly, researchers spend too much time preparing the data for analysis.
According to some estimates, data scientists devote 80% of their work to organising and cleaning data. It is clear that limited health data standardisation stalls research progress.
What are the benefits of standardising the health data?
Health data must be standardised and harmonised before researchers can collaborate quickly and effectively across global health data resources. Increased collaboration can lead to new insights and discoveries in the healthcare and research sector.
Below are further details on some key benefits gained when health data is standardised.
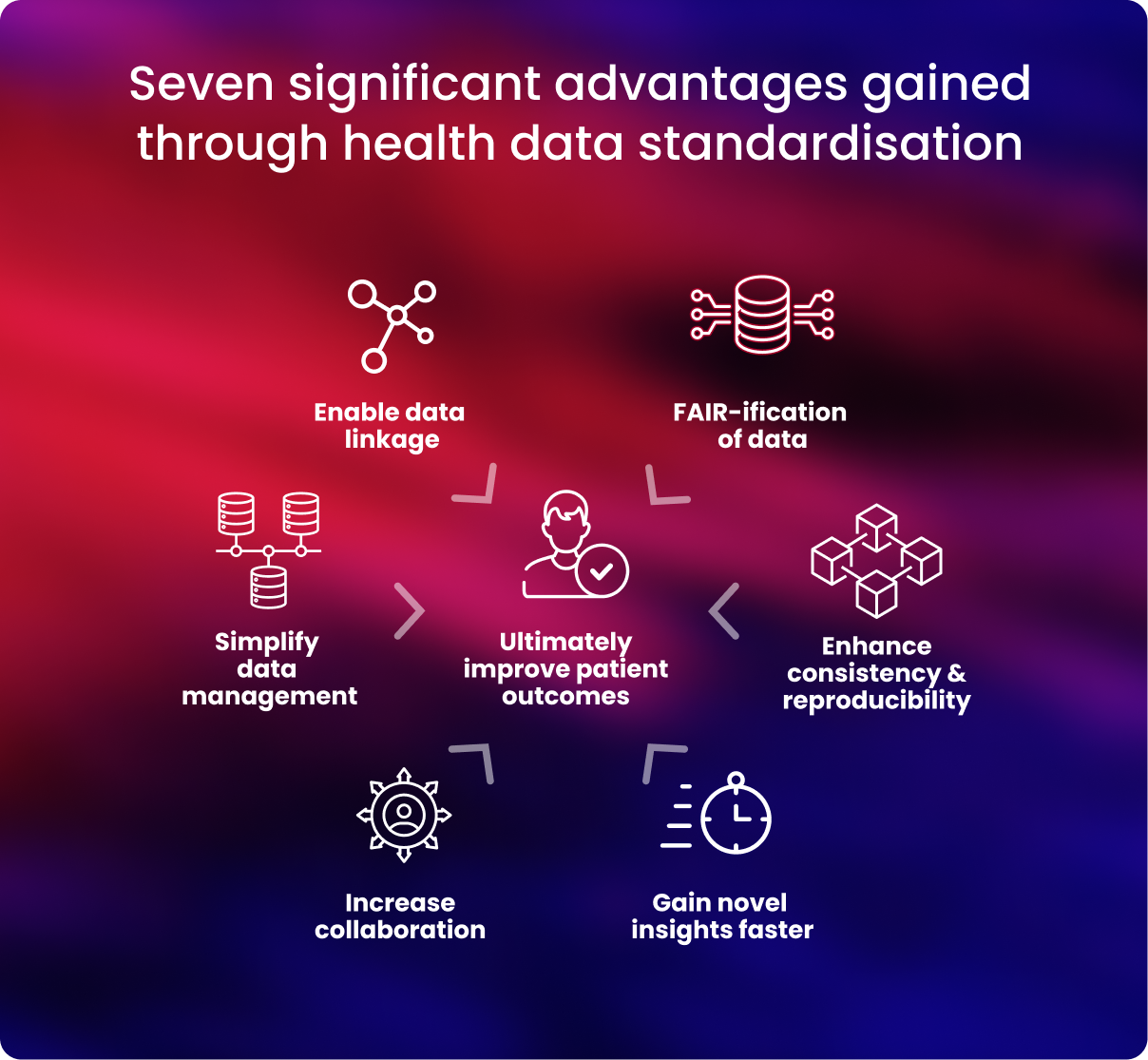
- Data management is streamlined
-
Researchers can establish data harmony by employing a single CDM. This facilitates uniform data storage in databases and frees up researchers' time for data analysis.4
- Standardised data is well aligned to the FAIR principles
-
To maximise research productivity, translate research, and advance precision medicine, health data transformation guarantees that data are compliant with the FAIR standards—data is findable, accessible, interoperable, and reusable.5, 6
- Data quality, consistency, and reproducibility are improved
-
Rigorous data quality is ensured, and problems with data integrity can be avoided by employing uniform data standards. Health data that has been standardised makes it much simpler to identify problems and ensure correctness, giving researchers access to accurate data for their analysis.
- Standardised health data can be robustly linked to other data
-
Medical data can be combined with other forms of data, such as lifestyle, environmental, or social data, to improve researchers’ and clinicians' understanding of a person's health.
However, if data cannot be successfully connected and merged to boost its statistical strength, researchers' capacity to derive novel insights from it is limited. CDMs are essential to guarantee that data is interoperable and can be linked appropriately
- Global collaboration across datasets is simplified
-
It is simpler to communicate and use data when working with other researchers and clinicians when health data is standardised. By harmonising their data, researchers may ensure their work complies with the strictest international standards and is compatible with other cohorts and organisations.
Providing a common data format (eg CSV file) and schema (eg “data should be split in 3 tables”) make data standardised, harmonised and predictable. This enables researchers to query datasets from diverse sources simultaneously, for example, a researcher could run a query to “Find all participants with disease X in biobank 1, biobank 2 and my private dataset.
Researchers are increasingly using trusted research environments (TREs) and federated analysis to safely access and evaluate standardised datasets to support worldwide collaboration fully. 8
Normalising all data to internationally recognised standards allows researchers to perform joint analyses across distributed datasets, which is key to ensuring diversity and representation of as many populations as possible in studies - Discoveries are accelerated
-
When health data is uniform, researchers may more easily combine, compare and study it concurrently to gain insights they can apply to further their understanding.
For example, one genome-wide association study showed that increasing sample size by 10-fold led to a 100-fold increase in findings, enabling genetic variants of interest to be more easily validated and studied.9
Additionally, when data is harmonised, it can be used for simplified, standardised and enhanced analytics, allowing researchers to transform data into discoveries. - Improve patient outcomes
-
Taken together, the advantages that data standardisation brings are that it becomes genuinely interoperable, shareable, and exploitable.
Researchers may statistically boost their research by integrating data for analysis, enabling quicker clinical applications and better patient results.
Enabling health data standardisation - technical challenges and solutions
While it is clear that many benefits can be gained by standardising health data to a common format, it is not without technical challenges. Below are some challenges researchers can face when standardising health data.

- Choosing the most suitable CDM
-
CHALLENGE
Different medical languages or ontologies may be favoured by organisations or researchers.
The type of data, how the data is being acquired (eg, through a clinical trial or observationally), the analysis it is being used for, and the simplicity and speed of usage required will all influence which CDM is most suitable.
SOLUTIONIn large-scale, international population-scale health investigations, OMOP is rising in favour as a regularly used CDM. Global pharmaceutical corporations, research organisations, and biobanks utilise OMOP to standardise clinical data. Therefore, when an organisation or researcher decides which CDM to use, it should be considered a strong front-runner. Examples of health organisations utilising OMOP as their CDM:
- The National Health Service (NHS) in the UK
- UK Biobank
- Genomics England
- All of Us from the National Institutes for Health (NIH) in the US.
- Inability to standardise, access, and securely analyse data.
-
CHALLENGE
health data users (64%) lack the knowledge to standardise data quickly. However, the standardisation of health data brings significant advantages. Furthermore, users can bring standardised analytical tools to the location where the data is stored in its secure environment once it has been standardised. To increase the amount of insights that can be obtained, secure access to and analysis of the data must also be harmonised.
SOLUTION
End-to-end solutions that securely standardise and provide health data access and analysis are essential.Researchers can simplify this procedure by using extraction, transformation, and loading pipelines (ETL) that can automate the standardisation process and transform unprocessed data into data ready for analysis.
Additionally, "no/low-code" tools are becoming more popular. Regardless of data science experience, more people can utilise these tools thanks to their intuitive design. This assists in democratising health data and the conclusions drawn from it.
An example of this is The Galaxy Community, an ELIXIR component. It offers a web platform for computational research in several ‘omics’ domains. 12
Another example is the NIH’s Common Fund Data Ecosystem (CFDE) Search Portal. The CFDE is a comprehensive resource for datasets generated through NIH funding, aiming to make data more usable and useful for researchers and clinicians. There are interactive search functions and visualisations of gene-specific, compound-specific, and disease-specific data and more to empower researchers with and without a data science background.
These technologies allow users of various backgrounds to access this health data directly or create complex workflows and replicable analytical pipelines. They also offer components of a truly end-to-end system, which could be integrated into a comprehensive solution in theory.
- Reduced data quality and loss
-
CHALLENGE
The data quality must not be compromised during the harmonisation of health data. Data quality is crucial to guarantee that reliable inferences may be formed from the data.
The primary healthcare environment, the validity and repeatability of study findings, and the potential usefulness of such data can all be negatively impacted by low-quality data.
SOLUTION
Following data transformation into a standard format, data quality must be evaluated. Along with data transformation, standardisation solutions should include stringent data quality checks.
With Lifebit's Data Transformation Suite, data is standardised, mapped to existing standards, annotations, and ontologies, and then connected together during data input to generate a linked data graph. Steps for quality assurance and control are also included, ensuring that data quality is maintained throughout the harmonisation process. - Standardisation is a time-consuming and laborious task
-
CHALLENGE
It takes time and effort to ensure that health data is harmonised.
Some estimates state that data scientists devote 80% of their work to organising and cleaning data.
SOLUTION
Technological advancements, such as ETL pipelines and end-to-end solutions, provide an optimal answer. These tools enable academics and medical professionals to focus time on what matters: developing new insights to improve patient outcomes.
The Data Transformation Suite from Lifebit employs several processes to convert unprocessed data into data suitable for analysis. Furthermore, these automated yet adaptable pipelines were designed to handle new data kinds as they emerge.
- Risks to security if data transformation is outsourced
-
CHALLENGE
Data standardisation and transformation can be assisted by third parties. However, since downloading the data puts it at risk for interception, this frequently compromises data security.
SOLUTION
Researchers can avoid the problems of decreased security and increasing expenses to standardise data by using federated technologies where experts standardise data but it never leaves its jurisdictional boundaries, researchers and clinicians avoid the issues of reduced security and increased costs to standardise data.
Key players driving health data standardisation
International initiatives are coming together to tackle the issue of limited health data collaboration and interoperability. Some of these are listed below:

Global Alliance for Genomics and Health (GA4GH)
A worldwide policy-framing alliance across 90 countries of various stakeholders, recognises the urgent need to enable broad data sharing across the borders of any single institution or country. Additionally, they highlight that researchers are effectively locking away the potential of data to contribute to research and medical advances by not doing this.

A worldwide policy-framing alliance across 90 countries of various stakeholders, recognises the urgent need to enable broad data sharing across the borders of any single institution or country. Additionally, they highlight that researchers are effectively locking away the potential of data to contribute to research and medical advances by not doing this.

Health Data Research UK (HDR UK) works to integrate health and care data from throughout the UK to allow advancements that enhance people's lives. They combine, enhance, and use healthcare data as a national institute.

The UK Health Data Research Alliance was formed to bring together leading healthcare and research organisations as an independent union to create best practices for the ethical use of UK health data for large-scale research.
![]()
The European Health Data and Evidence Network (EHDEN) has established a standardised ecosystem of data sources to reduce the time needed to provide answers in health research.

CDISC is an international, non-profit charity organisation that brings together a community of specialists from many fields to improve the development of data standards. They aim to make data more easily accessible, interoperable, and reusable for research so it can have a bigger impact on global health.

HL7 FHIR Foundation, an international, non-profit organisation. The foundation offers data, tools, and project support to aid in the collaboration, alignment, and growth of the FHIR community. To enhance healthcare quality, efficacy, and efficiency, the foundation works to make health data more interoperable.
Health data standardisation in action: enabling data linkage
Health data transformation has clear advantages, but what has it helped clinicians and researchers achieve so far?
Bigger, better, more complex datasets may be produced when several forms of health information on the same individual are standardised and then linked, such as when electronic primary care records are integrated with hospital mortality statistics.

Data linkage can involve bringing together multiple relevant datasets for the same individual. These datasets may have been collected in different formats but must all be standardised. Data linkage can offer a more complete picture of a person's health, allow for better clinical trial follow-up, and collect real-world data on whole populations to help shape health policy. This presents more opportunities for researchers to make novel discoveries and advance healthcare.
Data linkage, however, has its challenges. Patient trust and consent, as well as ensuring that data management and usage are properly regulated, standardised and of appropriate quality, are all potential areas that must be considered.
What has data linkage enabled researchers to discover?
Standardised, linked datasets can provide important insights and eventually enhance lives. As shown in the image above, these linked datasets help form a complete picture of a person’s health.
Recent groundbreaking studies that illustrate the power of big, standardised data in health research include
- research confirming that high blood pressure is a risk factor for dementia- here, the National Institutes of Health (NIHs) All of Us database of EHRs on +125,000 participants was utilised
- the 100,000 Genomes study on rare diseases.
- research reporting the host characteristics triggering severe COVID-19 on approximately 60,000 participants.

Featured resource: Read our blog on data linkage- Better together: the promise of health data linkage - and its challenges
Future outlook: What can be discovered when more health data is standardised and linked?
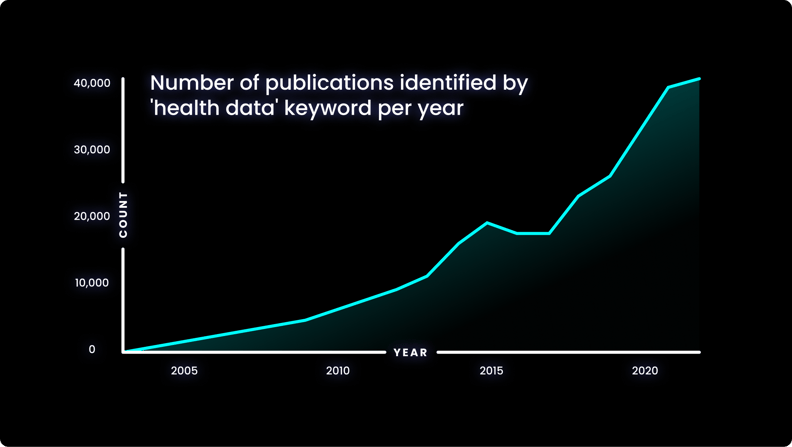
The increasing prevalence of health data in research is evident when considering the quantity of search results on PubMed for the terms ‘health data’- shown in the graph below.
The number of results for this term has increased significantly in recent years, and this is a trend that shows no sign of slowing down.
Globally, initiatives are investing in the continued collection, standardisation and analysis of linked health data to improve patient outcomes:
- In the UK, Our Future Health has begun linking multiple health sources and health-relevant information for up to five million participants. This will help create an incredibly detailed picture that reflects the whole of the UK population
From this, researchers will be able to make discoveries about both health and disease states, such as:
- generating individual risk scores for diseases – enabling precision medicine, better testing of emerging diagnostics and treatments.
- closing the gap in health inequality by including data more reflective of the diverse population of the UK.
- In the US, one million Americans are being asked to participate in the NIH’s All of Us Research Programme to create one of history’s most varied health datasets.
Researchers will use the information to understand better how biology, way of life, and environment impact health. They could use this to discover cures and illness prevention measures.
By enabling comprehensive, standardised data analysis, eradicating fragmented access to data and opening up secure access to distributed biomedical data, precision medicine approaches to health and disease can truly be realised.
In Brazil, Gen-t aims to combine biotechnology with genomic data from the country's population. Currently, 78% of all genomic data available for research comes from people with white European ancestry, while less than 1% t is from people of Latin American / Hispanic origin.
This imbalance significantly limits scientific insights’ impact on those in the region. To address the problem, Gen-T aims to involve 200,000 participants across five years and believes that increasing the variety of global genomic data will speed medical advances.
Health data standardisation: one part of the end-to-end data analysis pipeline
A key advantage of data transformation is that once data has been harmonised, users can bring industry-standard analysis tools to the secure environment where the data is stored.
However, to enhance the insights that may be acquired, the secure access to and analysis of the data should ideally be coordinated into a single solution. A comprehensive method of accessing, connecting, and analysing data while maintaining security is necessary to support data-driven research and innovation.
The advent of automated health data standardisation, trusted research environments and federated data analysis can provide the long-needed, comprehensive solution to balancing efficient data standardisation, access, analysis and security.
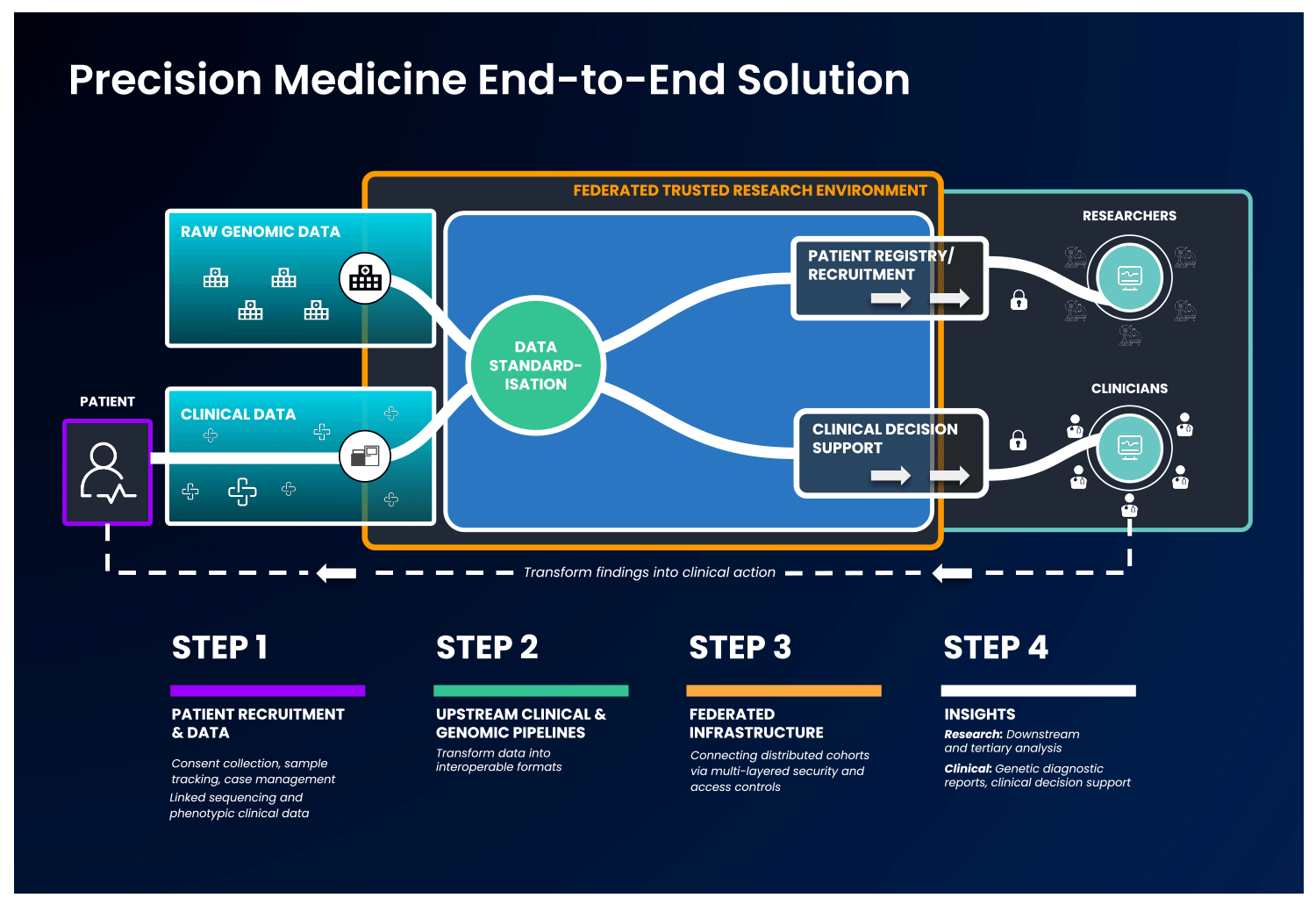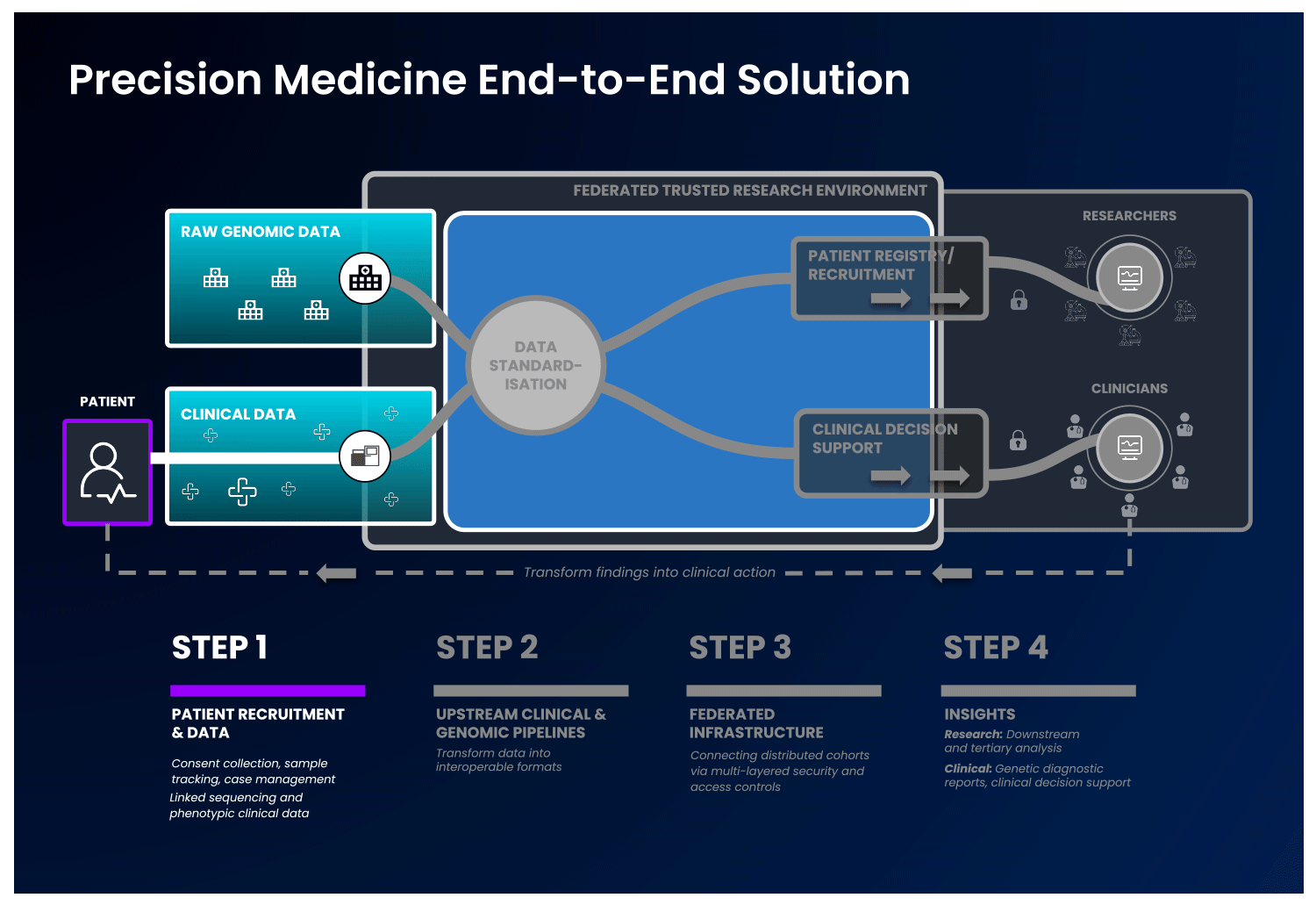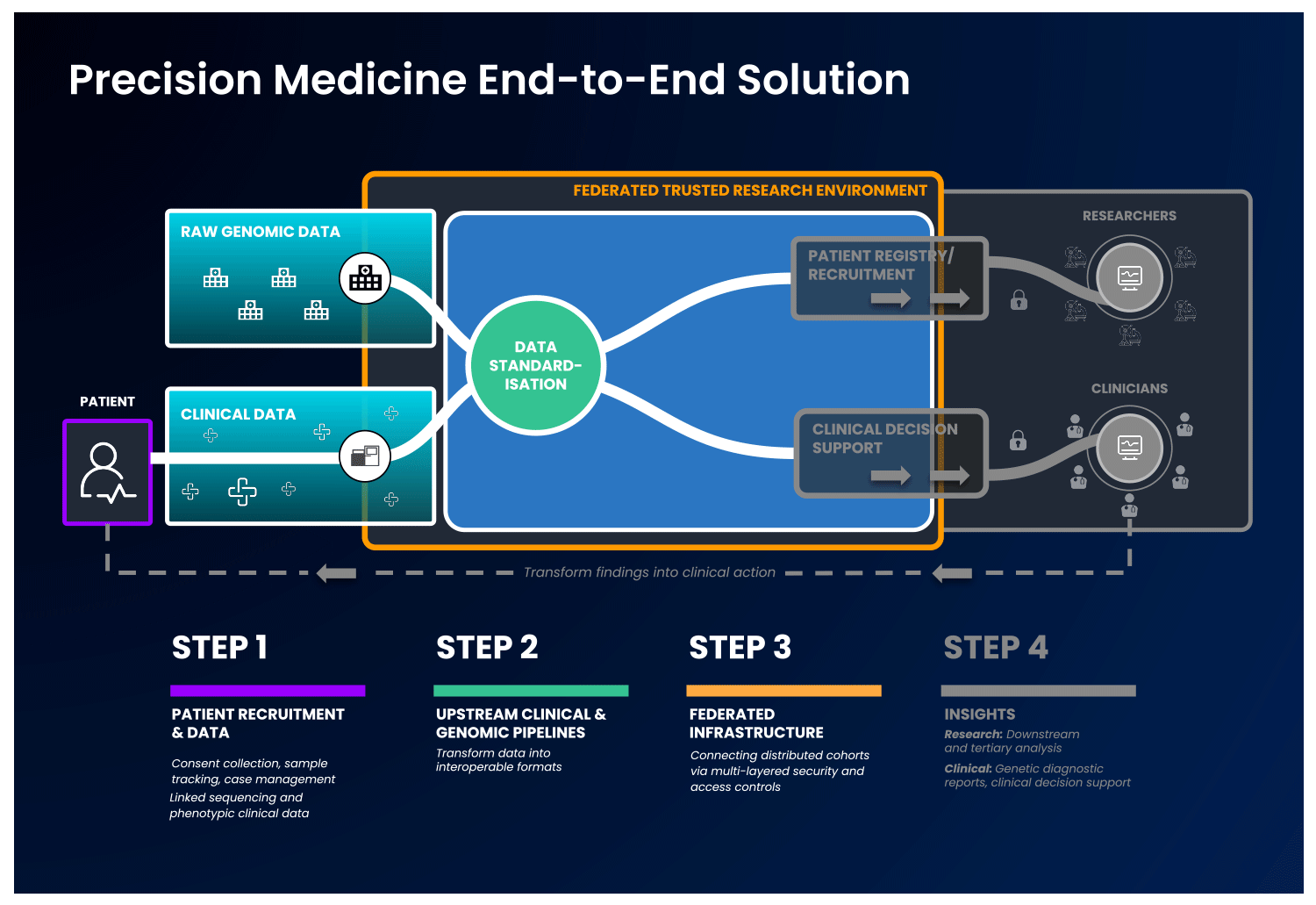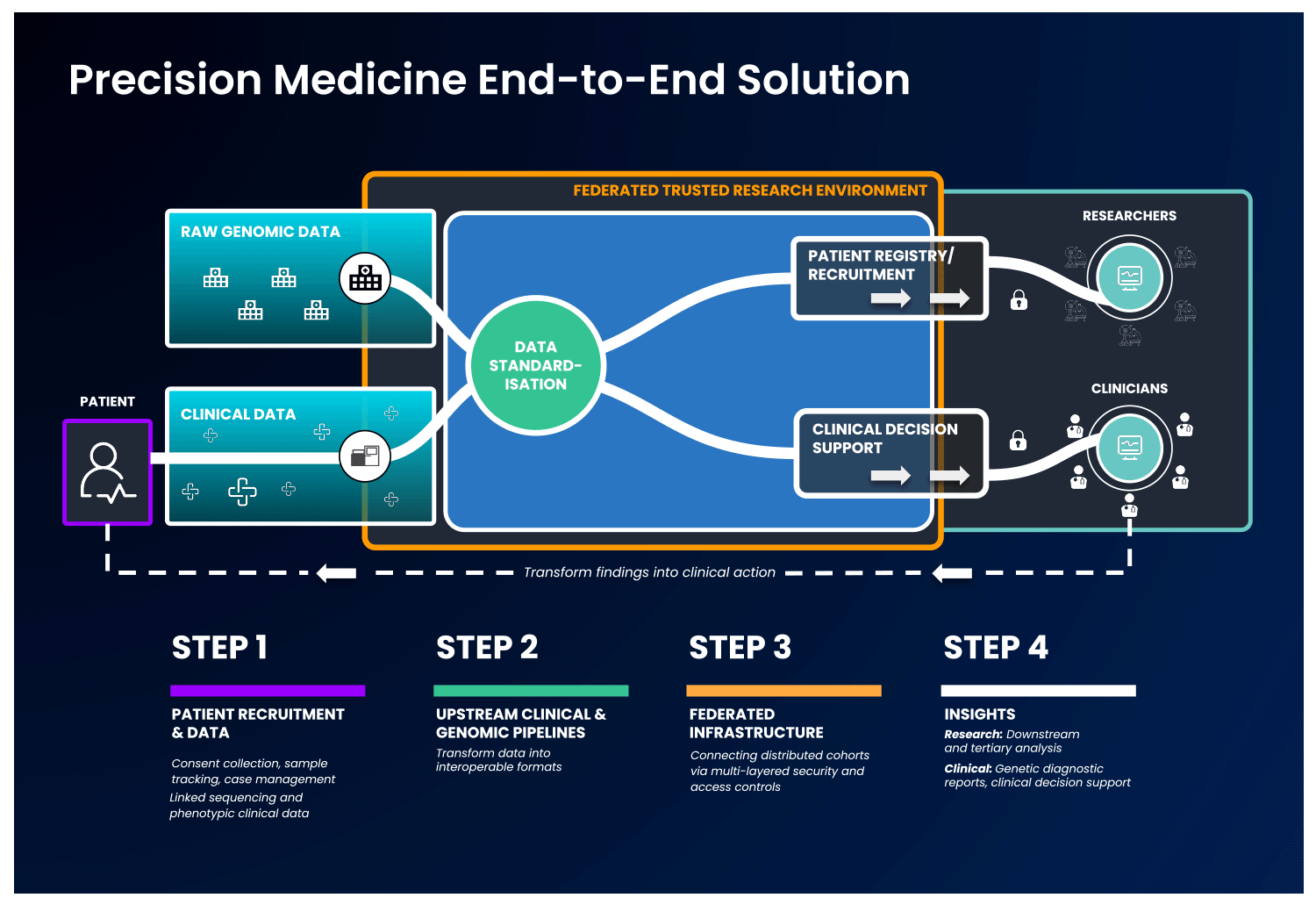
In an end-to-end, single solution, health data is first collected and standardised into interoperable formats. Next, these data are ingested into a cloud-based federated architecture, as described previously, to allow authorised users to access and combine this data with other disparate sources. With this, users can select unique and valuable cohorts of interest and perform downstream analysis without ever having to move the data, to benefit patients without compromising data security.
Strict security measures should govern the end-to-end solution. Security safeguards should be present at the levels of access, data and system security, with specific measures detailed in the image below:
It is possible to imagine an end-to-end platform that could securely integrate a country’s healthcare network, national genomic medicine initiatives and sequencing laboratories, progressing therapeutic discovery while keeping the data safe.
By taking full advantage of advances in data standardisation, cloud computing, federated data analysis and end-to-end data management platforms, research institutions, healthcare systems, and genomic medicine programs globally can harness the benefits of collaboration and joint analyses.
Finally, connecting diverse datasets can also help democratise access to data and insights while ensuring organisations retain autonomy over data.

Featured resource: Catch up on our data diversity webinar where we were joined by Prof Lygia V. Pereira, CEO and Co-Founder, gen-t Science, Victor Angel-Mosti, CEO and Founder of Omica.bio to discuss the challenges and opportunities surrounding health equity.
End-to-end, low-code platforms are democratising access and providing new perspectives on clinical-genomic data, advancing research and leading to significant therapeutic discoveries. These efforts will facilitate benefits sharing and promote equitable access to data and clinical insights and international scientific collaboration.
Summary
Health information exists in a wide variety of formats and sources. Data can only be properly integrated to provide fresh insights when standardised and interoperable.
Standardising health datasets is crucial to guarantee data quality and promote collaboration in healthcare and research. This process, combined with end-to-end data access and analysis solutions, is crucial to standardise, store and integrate healthcare data to deliver actionable insights.
Author: Hannah Gaimster, PhD
Contributors: Hadley E. Sheppard, PhD and Amanda White
About Lifebit
Lifebit provides health data standardisation, TREs and federated data analysis for clients, including Genomics England, Boehringer Ingelheim, Flatiron Health and more, to help researchers transform data into discoveries.
Lifebit’s services are making health data usable quickly.
Interested in learning more about Lifebit’s health data standardisation services and how we accelerate research insights for academia, healthcare and pharmaceutical companies worldwide?
References
- Papez, V. et al. Transforming and evaluating the UK Biobank to the OMOP Common Data Model for COVID-19 research and beyond. J. Am. Med. Inform. Assoc. 30, 103–111 (2023).
- Hume, S., Aerts, J., Sarnikar, S. & Huser, V. Current applications and future directions for the CDISC Operational Data Model standard: A methodological review. J. Biomed. Inform. 60, 352–362 (2016).
- Vorisek, C. N. et al. Fast Healthcare Interoperability Resources (FHIR) for Interoperability in Health Research: Systematic Review. JMIR Med Inf. 10, e35724 (2022).
- Junkai Lai et al. Existing barriers and recommendations of real-world data standardisation for clinical research in China: a qualitative study. BMJ Open 12, e059029 (2022).
- Wilkinson, M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
- Vesteghem, C. et al. Implementing the FAIR Data Principles in precision oncology: review of supporting initiatives. Brief. Bioinform. 21, 936–945 (2020).
- Mayo, K. R. et al. The All of Us Data and Research Center: Creating a Secure, Scalable, and Sustainable Ecosystem for Biomedical Research. Annu. Rev. Biomed. Data Sci. 6, 443–464 (2023).
- Alvarellos, M. et al. Democratizing clinical-genomic data: How federated platforms can promote benefits sharing in genomics. Front. Genet. 13, (2023).
- Visscher, P. M. et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 101, 5–22 (2017).
- Grannis, S. J. et al. Evaluating the effect of data standardization and validation on patient matching accuracy. J. Am. Med. Inform. Assoc. 26, 447–456 (2019).
- Schneeweiss, S., Brown, J. S., Bate, A., Trifirò, G. & Bartels, D. B. Choosing Among Common Data Models for Real-World Data Analyses Fit for Making Decisions About the Effectiveness of Medical Products. Clin. Pharmacol. Ther. 107, 827–833 (2020).
- The Galaxy Community. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50, W345–W351 (2022).
- Ehsani-Moghaddam, B., Martin, K. & Queenan, J. A. Data quality in healthcare: A report of practical experience with the Canadian Primary Care Sentinel Surveillance Network data. Health Inf. Manag. J. 50, 88–92 (2021).
- Rehm, H. L. et al. GA4GH: International policies and standards for data sharing across genomic research and healthcare. Cell Genomics 1, 100029 (2021).
- Nagar, S. D. et al. Investigation of hypertension and type 2 diabetes as risk factors for dementia in the All of Us cohort. Sci. Rep. 12, 19797 (2022).
- 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care — Preliminary Report. N. Engl. J. Med. 385, 1868–1880 (2021).
- Kousathanas, A. et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 607, 97–103 (2022).
- Sirugo, G., Williams, S. M. & Tishkoff, S. A. The Missing Diversity in Human Genetic Studies. Cell 177, 26–31 (2019).