Festival of Genomics 2019
.webp)
Lifebit
The Festival of Genomics has moved from the cavernous Excel stadium to the more intimate environment of the Business Development Centre in Islington this year, with a wide range of speakers from research, healthcare and industry. Here are a few of our highlights from the meeting…
Professor Mark Caulfield (Genomics England) started the session with an overview of the 100,000 Genomes Project and the work being done at Genomics England. Much had been reported previously, but one of the new points highlighted was around planned incorporation of pharmacogenomics in the Genomic Medicine initiative, citing an example from CPIC (Clinical Pharmacogenetics Implementation Consortium) in the USA, who had carried out analysis of 6,628 genomes for known pharmacogenomic markers, and identified at least one in 85% of all the genomes. Pilot reporting will be rolled out this year, and work is underway on how to share this information with patients so they can share with their doctors to aid better prescribing of drugs based on your personal genome data.
Dr Stefan Kirov described how Bristol-Myers Squibb applies NGS technology and bioinformatics to enhance drug discovery and development. Between 2011-2015, BSMS had generated 70 Tb of raw NGS data, which led to a need for hybrid cloud approaches for data storage and analysis. The pipelines that have been generated for DNA-seq and RNA-seq analyses have been optimised with enhanced QC steps at the beginning using their CohortMatcher tools, and this, along with other pipelines they use for CRISPR analysis, have been made openly available.
Dr Dennis Wang (University of Sheffield) described the breadth of work they have been doing within their department on standardising integration and analysis of healthcare data, and the outreach work through the Northern BUG (Bioinformatic User Group), to create standards used across more sites…

Dr Mohammed Kamran (Future Genetics) discussed the issue of equality of access to healthcare and the issues of representation of ethnically diverse clinical testing populations, including an example from 2015 where >31 cancer drugs were licensed by the FDA – less than 24 of these trials had <5% African American participants, while the incidence of cancer in 18 of the 24 drugs had the same or higher incidence in African Americans. Future Genetics is working to increase the ethnic diversity of clinical trial populations.
Applications of Human Genetics in Pharma Panel ( Dr Patrick Descombes, Nestle – Dr Nicola Beer, Novo Nordisk – Dr Victor Neduva, GSK). There were many questions from the audience including how to handle metadata and what would the audience do with access to the full 100,000 Genomes Project data. Metadata was acknowledged as a key issue, and some organisations have dedicated whole departments to the task. All panel members discussed following the 5R’s methodology from AstraZeneca – Right target, Right tissue, Right safety, Right patients, Right commercial potential (Nature Reviews Drug Discovery volume13, pages 419–431, 2014). More data is generally better, but genomic data should be used as a hook upon which to hang as many other data types as possible about all potential targets. Despite the vast amount of genomic data, it is still hard to identify druggable targets. Also, today’s data sets are predominantly based on small variant detection. New sequencing technologies, such as long reads, are starting to show us to detect larger structural changes and their role.
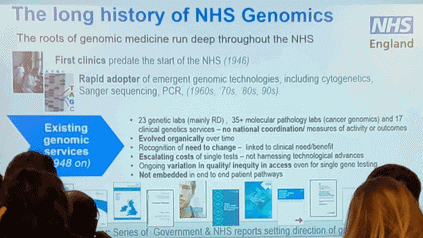
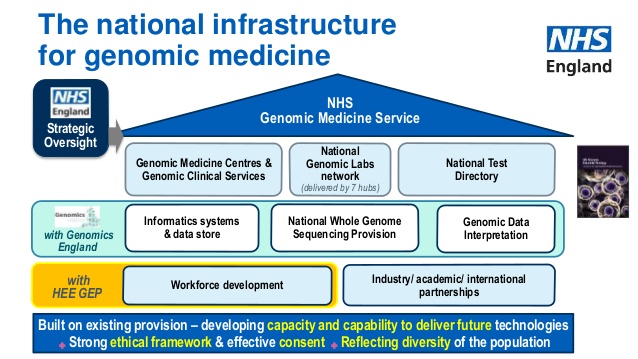
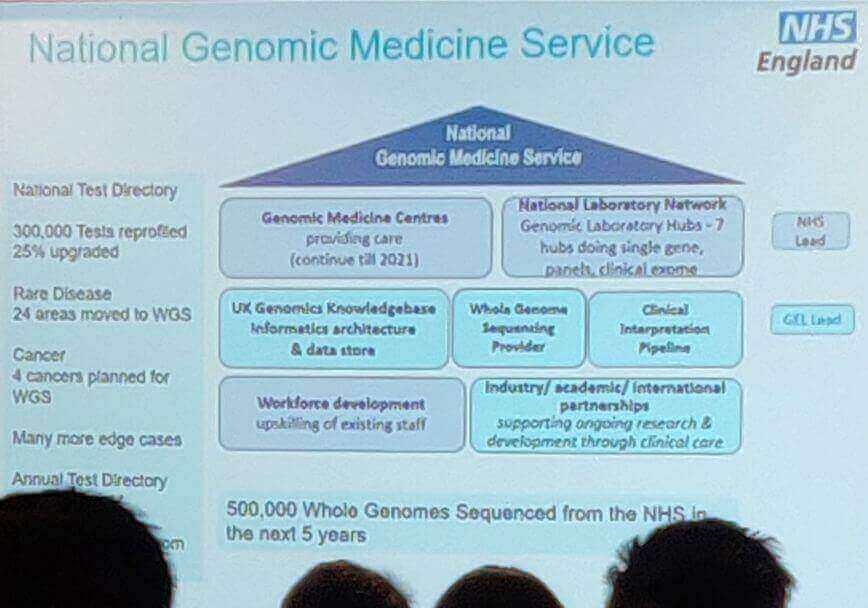
Professor Sue Hill (CSO, NHS) provided an excellent overview of the new Genomic Medicine Initiative in the NHS, and its historical context.
Dr Suzanne Brewerton provided an overview of AstraZeneca’s strategy towards applying genomic data for drug discovery and development. There is a company-wide drive to harness all data collected to improve the delivery of new drugs.
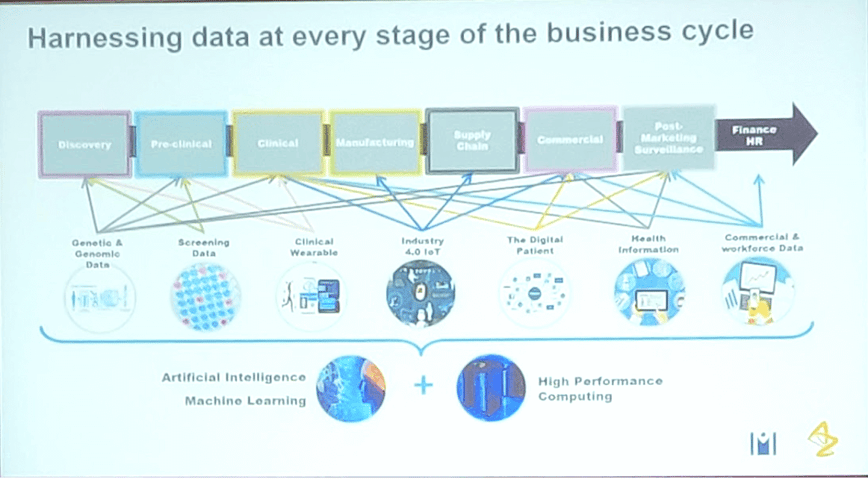
The path forward will involve bringing together disparate datasets, either physically or virtually, and linking them in different ways to address specific questions. With plans to sequence 2,000,000 genomes by 2025, there are a lot of both logistical and analytical issues to address.
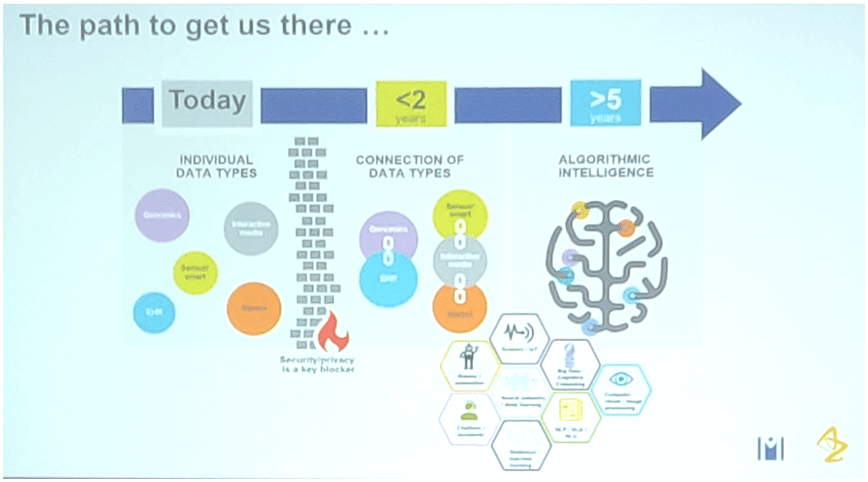
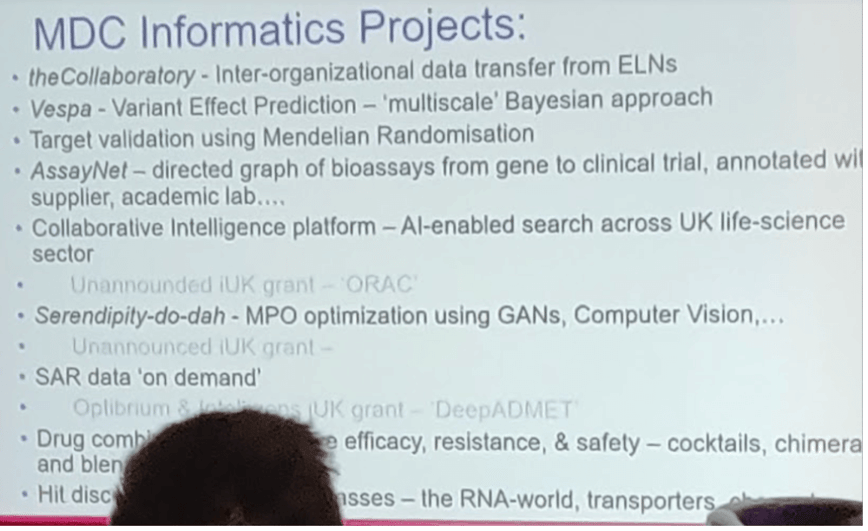
Professor John Overington described work at the Medicines Discovery Catapult of use informatics to bring together disparate silos of data (a theme running throughout the meeting). Providing examples of the resources that can then be utilised with SME’s and research groups to speed up new therapeutic agents, along with some interesting discussion on Blakes Seven (for the oldies in the audience), cocktails and malt whiskies…. as analogies, of course!
Dr Guillermo del Angel told us how Alexion Pharmaceuticals were approaching the challenges of large genomics data sets and integrating them with clinical data for better insights, both by going beyond the genome and integrating existing data and applying new technologies.
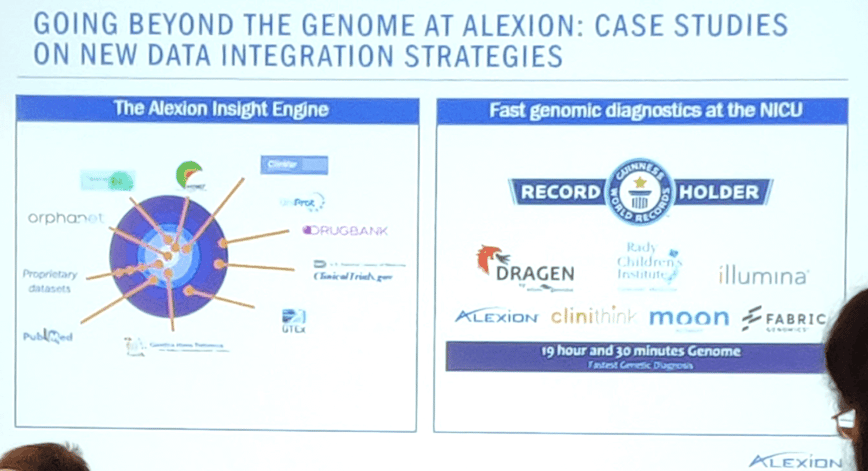
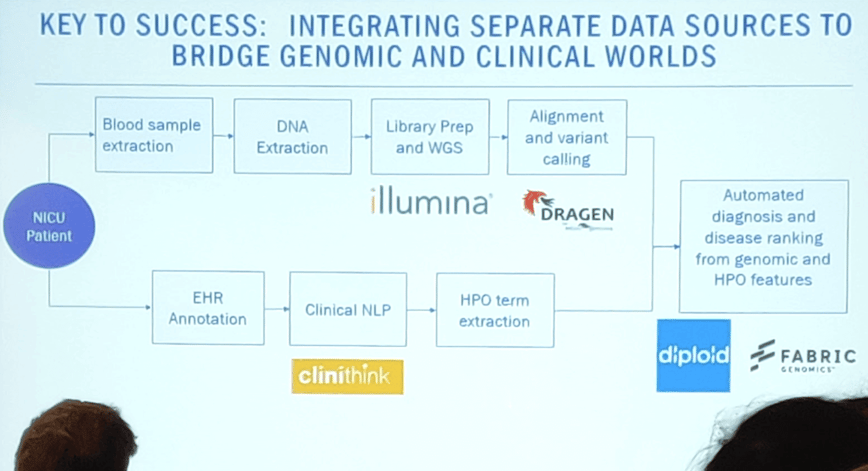
Alexion’s focus on treatments for rare diseases led to collaborations with the Rady Children’s Institute, Guinness World record holders of the fastest genomic diagnosis in 19.5 hours. This project brought together technology providers Illumina and Dragen, clinical tools from Alexion and Clinisight, and diagnostic interpretation from the Rady, Fabric Genomics and Diploid’s Moon platform.
Until next year, #genomicsfest!
We would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!