

Week 4: Calling All Researchers to Fight COVID-19 with Open Science
Lifebit
It’s been just 5 weeks since COVID-19 was declared a pandemic by the World Health Organisation (on March 11th to be exact) and the initiatives being launched throughout our research community continue to multiply… Welcome to our 4th installment of Lifebit’s curated COVID-19 Resources to help keep you up-to-date on these fast-moving developments, so that together we can all contribute our time and knowledge in this global fight.
Data Resources
CoVex
Drug repurposing is an attractive approach during the COVID-19 pandemic as it offers new therapeutic options through the identification of alternative uses for already approved drugs. Drug repurposing in times of crisis such as these, allows us to save precious time by working with what we already know and have available to us as a scientific community.
CoVex is a network and systems medicine online data analysis platform that integrates virus-human interaction data for SARS-CoV-2 and SARS-CoV. CoVex provides a user-friendly interface and integrates virus-host protein interactions of SARS-CoV-2 and SARS-CoV-1, the human protein-protein interaction network, and a comprehensive drug-target protein network. CoVex hence offers a systems-level approach to identify candidates for drug repurposing against SARS-CoV-2.
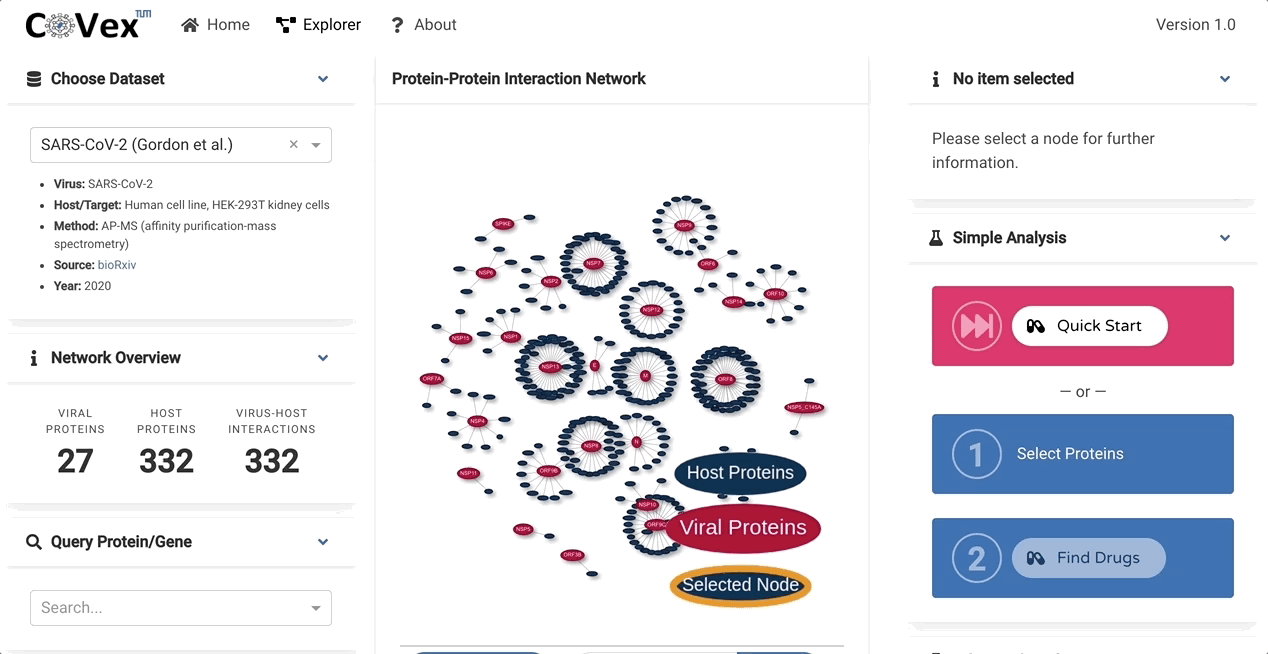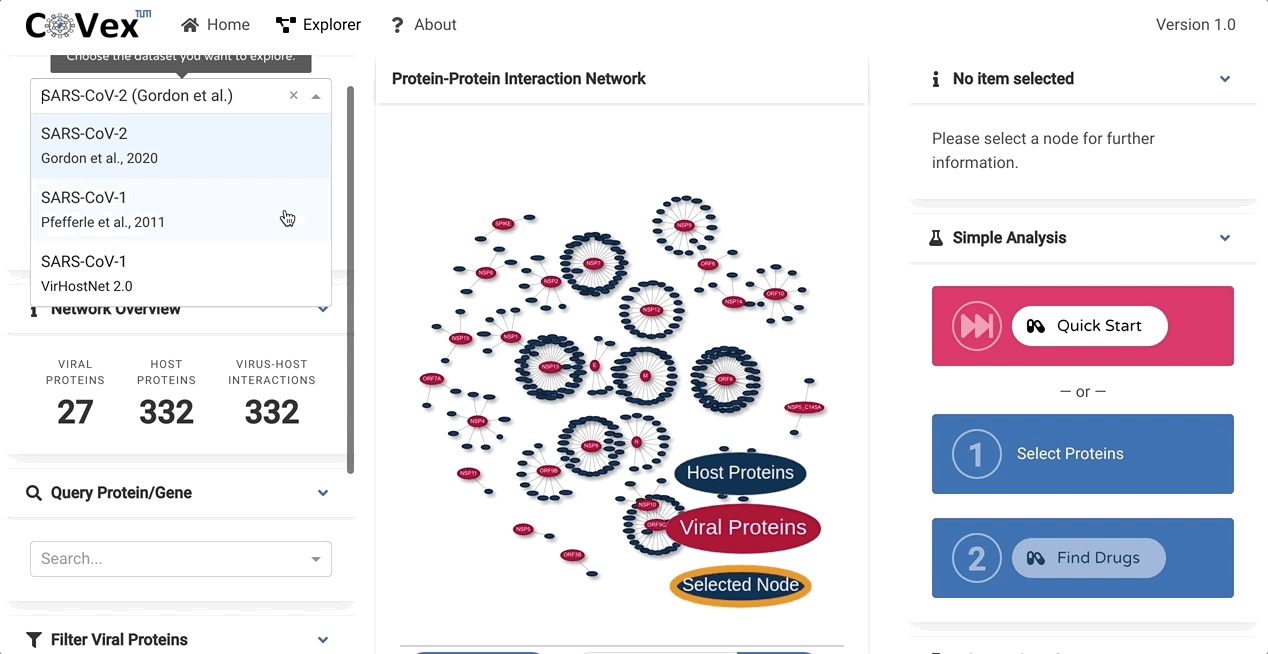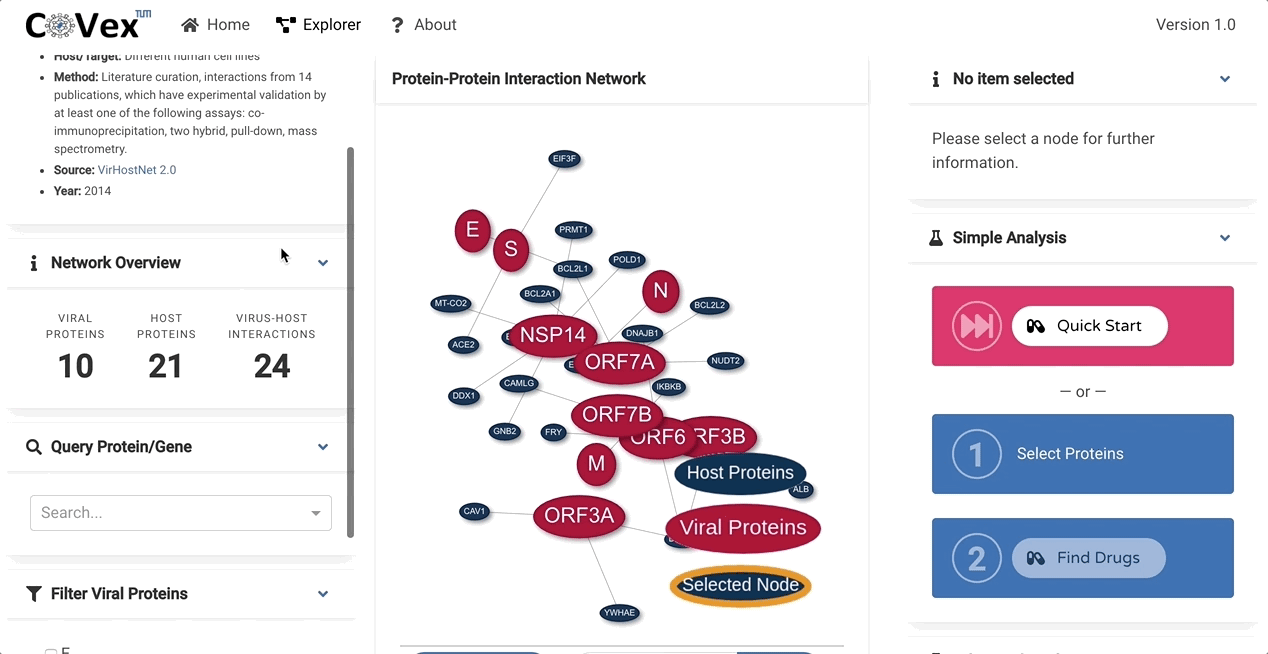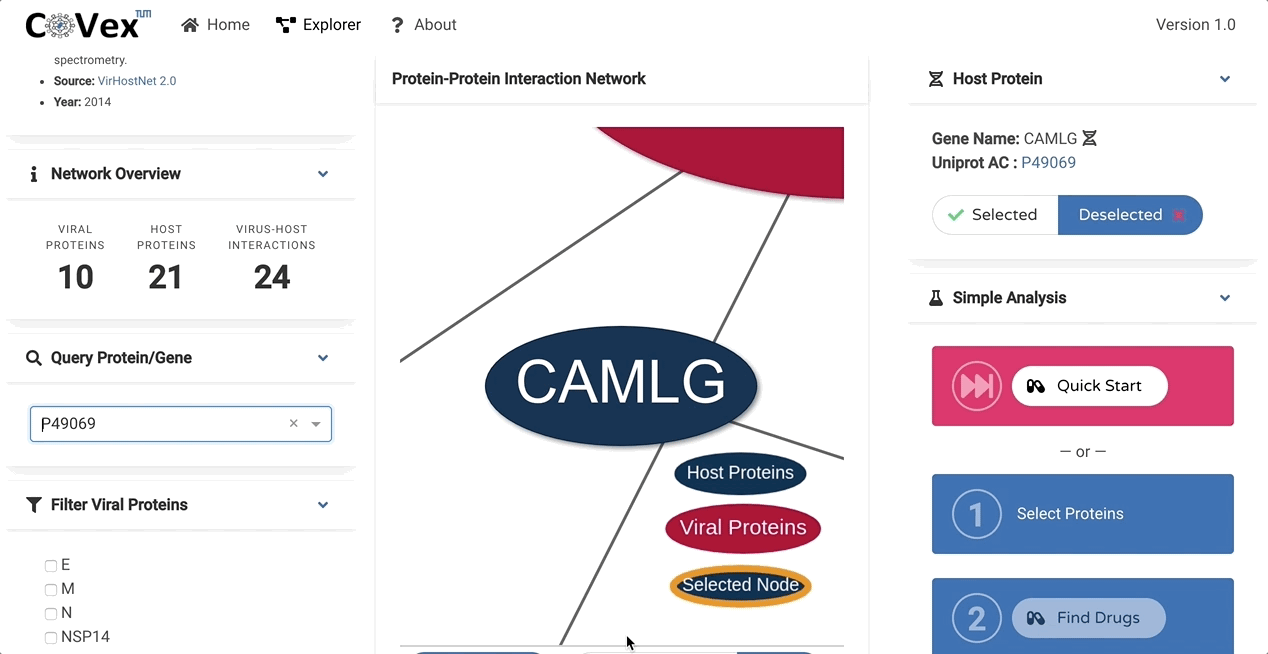
The main goal behind CoVex is making network and system medicine approaches, and their direct application to relevant data, available and easily accessible for a broad scientific audience.
CoVex has been developed by a team of researchers from the Chair of Experimental Bioinformatics (ExBio) at the Technical University of Munich (TUM).
Centre of Genomic Regulation (CRG): Uniform analysis of coronavirus and SARS-COV-2 nanopore direct RNA sequencing datasets
This resource is a joint initiative built by the Epitranscriptomics and RNA Dynamics Lab (Novoa Lab) and the Bioinformatics Core Facility (BioCore) at the Centre for Genomic Regulation (CRG), in Barcelona, Spain.
The main objective of this collaboration is to facilitate the community’s access to uniformly processed SARS-CoV-2 nanopore direct RNA sequencing datasets. As such, the team has processed all publicly available dRNAseq SARS-CoV-2 datasets using the MasterOfPores workflow.
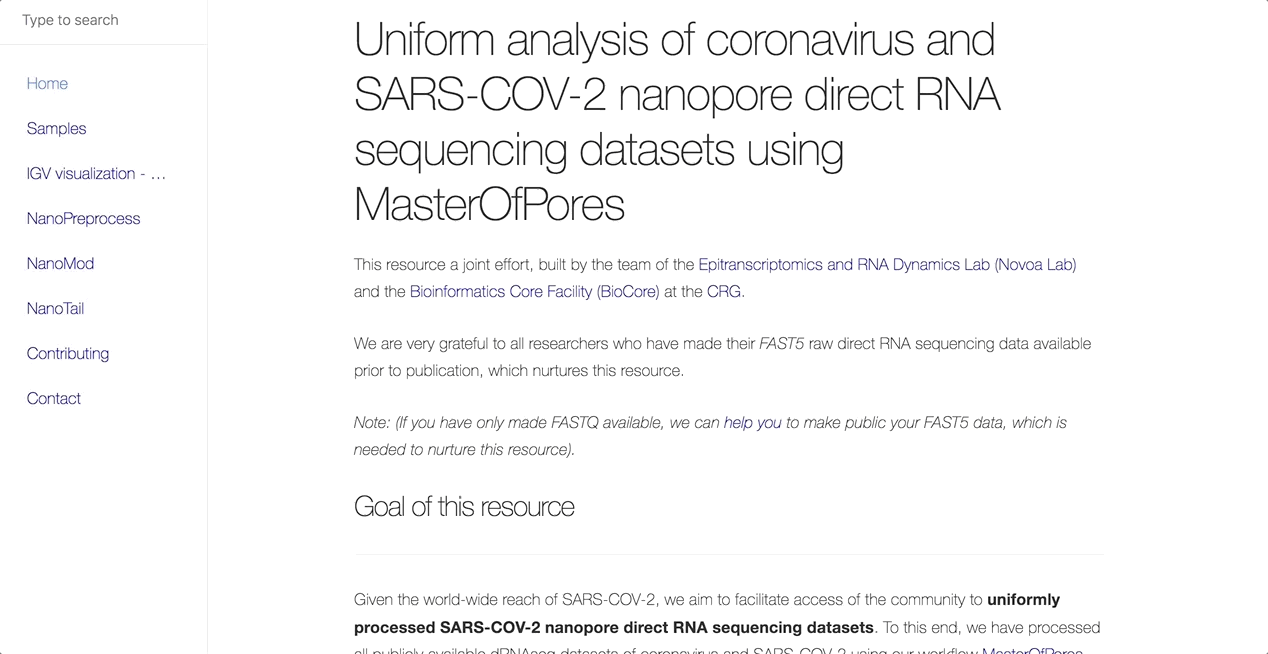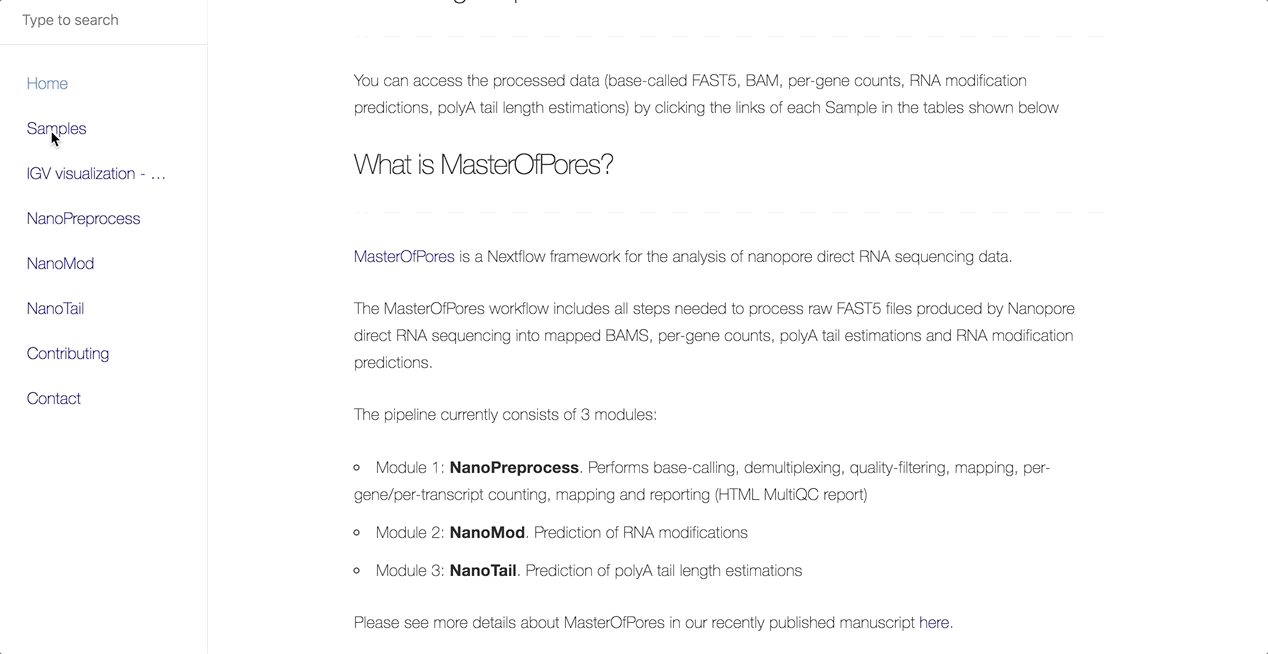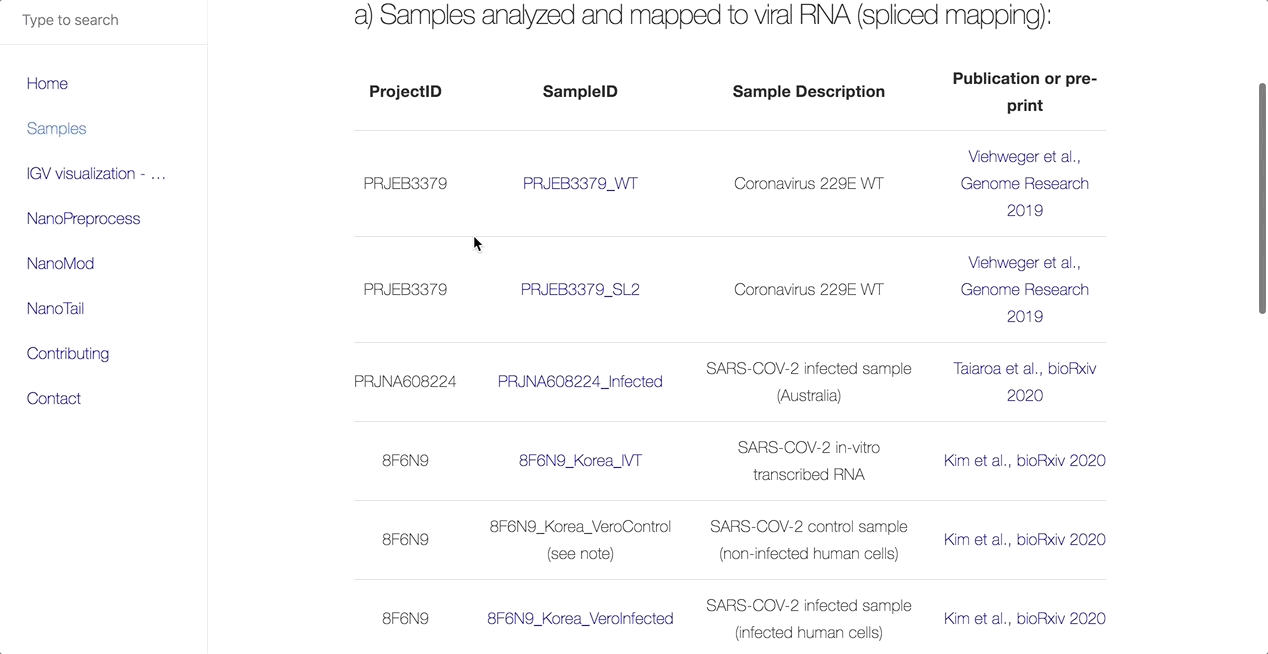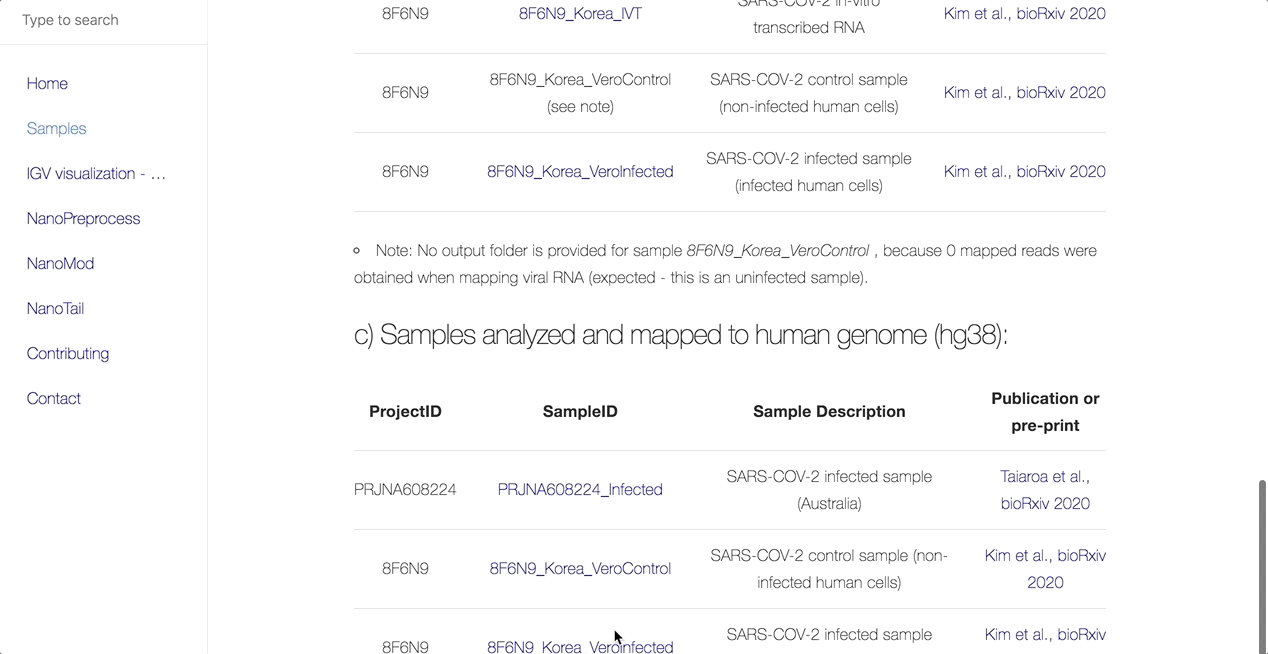
MasterOfPores (Cozzuto et al, 2020) is a Nextflow framework for the analysis of nanopore direct RNA sequencing data. It includes all steps needed to process raw FAST5 files produced by Nanopore direct RNA sequencing into mapped BAMS, per-gene counts, polyA tail estimations and RNA modification predictions.
COVID-19 Research Communities and Challenges
[COVID-19 hg] COVID-19 Host Genetics Initiative
The COVID-19 Host Genetics Initiative’s mission is to bring together the human genetics community to generate, share and analyse data to learn the genetic determinants of COVID-19 susceptibility, severity and outcomes.
Discoveries stemming from this initiative could help generate hypotheses for drug repurposing, identify individuals that have unusually high/low risk and contribute to global knowledge of the biology of SARS-CoV-2 infection and disease.
The initiative also includes a useful partner portal which allows researchers to explore studies and research questions, ultimately leading to global collaborations.
As part of the initiative, analyses have been made available to researchers to better understand the genetic basis of COVID-19 susceptibility and severity, which can be easily accessed through this GitHub repository.
COVID-19 HPC Consortium
Fighting COVID-19 requires extensive research in areas like bioinformatics, epidemiology, and molecular modeling to understand the threat we’re facing and create strategies to address it – all of which requires a massive amount of computational capacity.
The COVID-19 HPC Consortium is a collaboration of over 30 supercomputers that harness over 400 petaflops of computing power, affording researchers plenty of supercomputing capacity to bring us one step closer to defeating the novel coronavirus.
The compute resources are offered free of charge to researchers around the world in order to boost our chances of finding approaches to develop drugs and vaccines to combat this new disease that has brought havoc to the world.
Researchers are encouraged to submit COVID-19 related research proposals via the online portal, which will then be reviewed for matching with resources from one of the Consortium partners. A special emphasis will be placed on projects that can ensure rapid results.
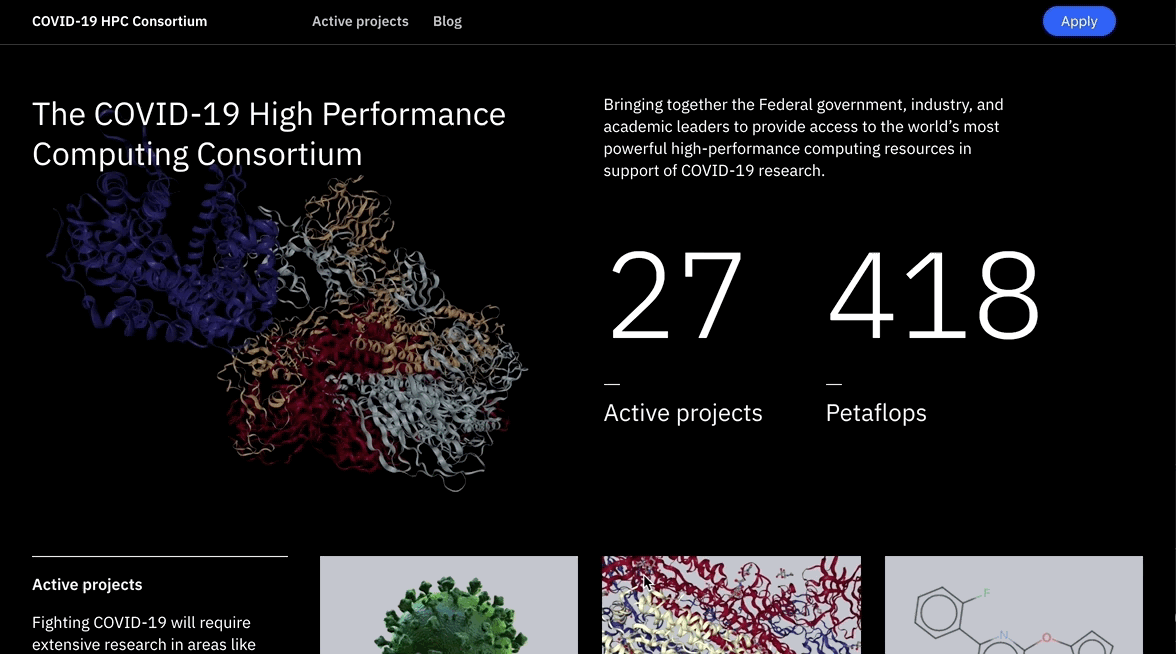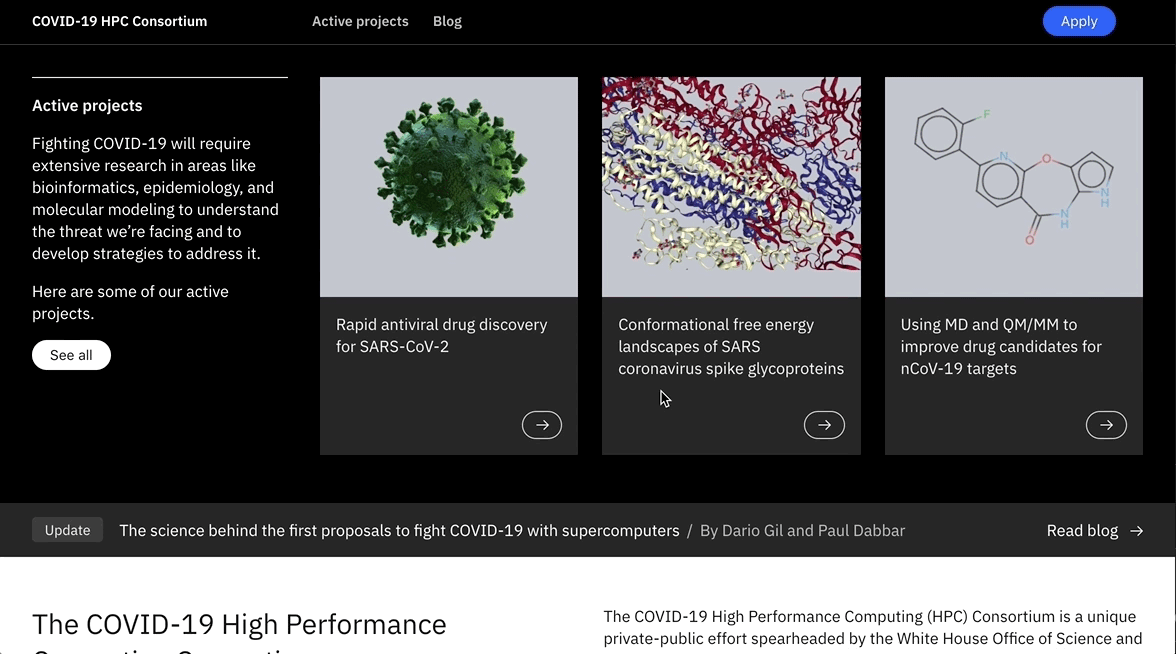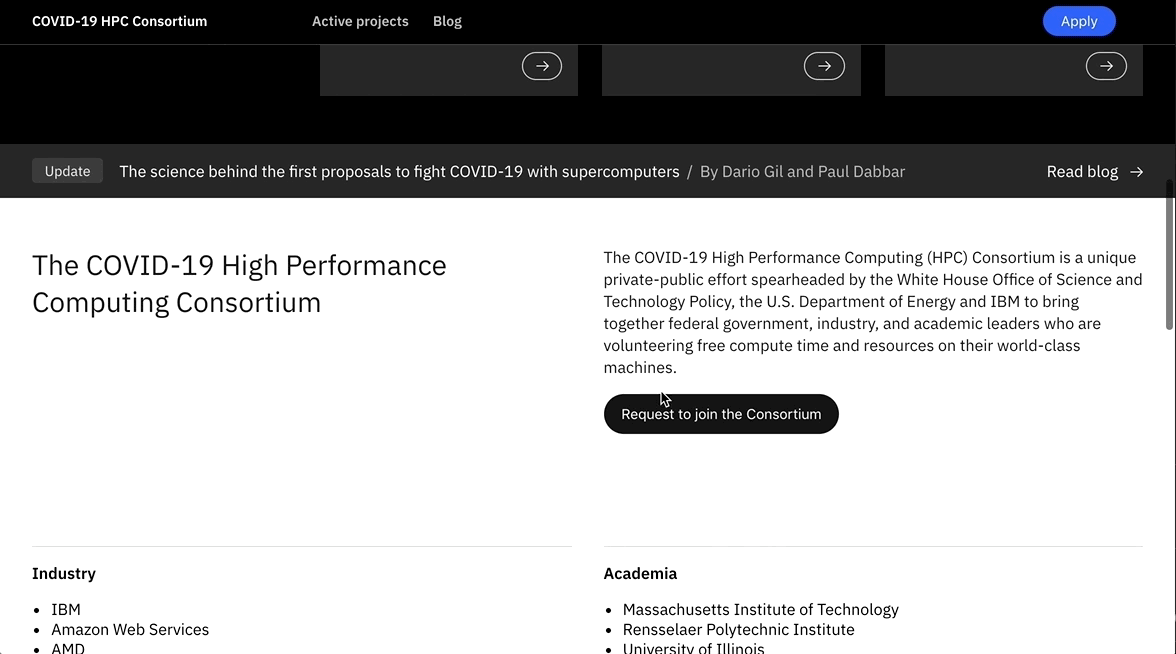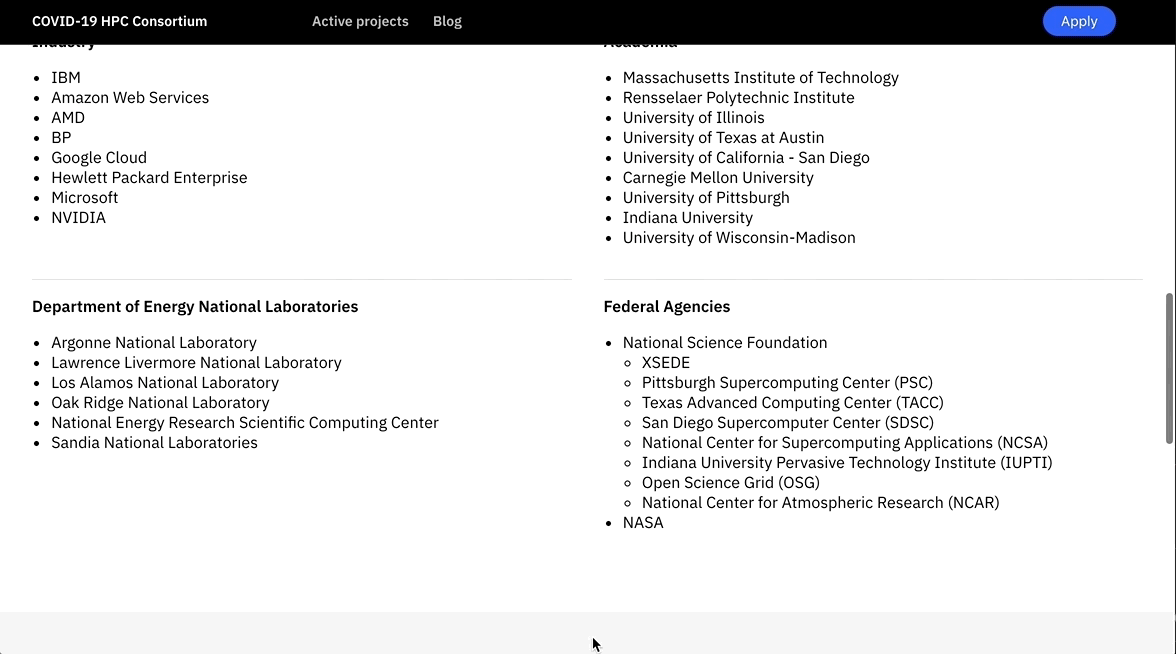
The COVID-19 HPC Consortium, co-lead by the U.S Department of Energy and IBM, brings together government, academia, and industry. Providers include Google, Microsoft, Amazon, Hewlett Packard Enterprise, Nvidia, AMD, NASA, the National Science Foundation, the Pittsburgh Supercomputing Center and the Wyoming Supercomputing Center. Six U.S. National Labs are also on board – Lawrence Livermore, Lawrence Berkeley, Argonne, Los Alamos, Oak Ridge and Sandia, as well as academia, including MIT, Rensselaer Polytechnic Institute, University of Illinois, University of Texas at Austin, University of California – San Diego, Carnegie Mellon University, University of Pittsburgh, Indiana University, and the University of Wisconsin-Madison.
Let’s keep the conversation going – please share with us any COVID-19 resources that you have found valuable. We will continue to update our readers and followers regularly. We are in this together!
Featured news and events
2025-03-26 11:17:46
2025-03-14 15:45:18
2025-03-05 12:49:53
2025-02-27 10:00:00

2025-02-19 13:30:24
2025-01-30 12:47:38

.png)