

How-To Guide: Standardised Secondary Genomics Analysis with Illumina's DRAGEN
Lifebit
The quest for robust, rapid, scalable & integrated genomics analysis
In today’s world of bioinformatics, data has gotten too broad and complicated, creating intricate data silos which impede researchers from unlocking the true value of data. Systems can no longer cope, and instead, turn into hurdles that give rise to scalability, management and maintenance issues. Integrated Analysis (IA) is a holistic, FAIR and federated approach to data analysis. It requires uniting disconnected, diverse data and transcending system, silo and collaboration barriers, and results in impactful insights.
A central tenet of the Integrated Analysis methodology and bioinformatics, in general, is data standardisation which enables researchers to transform raw siloed data into impactful insights. Every robust, multi-step bioinformatics workflow includes three analytical stages: primary, secondary and tertiary analysis.
- Primary Analysis (sequence generation & quality control/assurance)
- Secondary Analysis (sequence processing)
- Tertiary Analysis (results interpretation)
The challenges of standardising & scaling secondary analysis
The lack of standardisation for secondary analysis is widely recognised in the field and can be attributed to the massive and overwhelming amount of constantly-evolving open-source solutions available to researchers. As software development for omics analyses is evolving at an increasingly fast pace (over 3,000 bioinformatics tools were developed in 2018 alone), standardisation becomes of utmost importance to ensure easily reproducible analyses and harmonisation. Furthermore, bioinformatic software solutions are often implemented as single-purpose packages, but can be strung together in order to develop a custom/ad-hoc secondary analysis pipeline. Stringing software manually, however, is an extremely tedious task requiring a lot of work and time. Lastly, secondary analysis is quite resource-intensive as it runs a set of time-consuming algorithms on a per sample basis.
When it comes to genome analysis, the BWA-Genome Analysis Toolkit (BWA-GATK), part of the Broad Institute’s Best Practices analysis pipeline, allows researchers to map and align reads against reference genomes, and perform variant discovery analysis from quality-scored sequence data derived from primary analysis. This is, by far, the most widely used analysis pipeline, enabling researchers to go from raw BAM files to VCF files.
BWA-GATK is not a cloud-native application, resulting in a slow, memory-intensive and unscalable pipeline. The end-user is directly responsible for optimising system utilisation and improving the scalability of the process.
Illumina’s DRAGEN: an ultra-rapid cloud-native implementation of BWA-GATK
Illumina’s DRAGEN is an accelerated and improved cloud-native implementation of the BWA/GATK standard (it can be accessed and run directly on AWS cloud through the AWS Marketplace). It resolves the issue of lengthy compute times for the secondary analysis of NGS data, allowing users to perform the secondary analysis of a whole human genome with 30x coverage in 25 minutes on-premise, which would have previously taken close to 15h with a traditional CPU-based system. When running secondary analysis on the AWS cloud, DRAGEN provides the same speed and accuracy as running on-premise, while also delivering the flexibility and scalability of the cloud.
Besides speed, Illumina’s DRAGEN has also proven itself to be highly sensitive and specific for detecting small variants (named in silico Variant Catcher of the PrecisionFDA Hidden Treasures 2018), leading to its implementation for ultra-rapid secondary analysis by some of the biggest names in genomics research, including Genomics England.
Illumina’s DRAGEN analyses data from different sequencing experiments such as whole genomes, targeted panels, germline/somatic datasets and RNA sequencing experiments. It can also be implemented in a variety of research applications including population sequencing, newborn intensive care units for rapid genomic testing, and clinical and translational research, among others.
Although Illumina’s DRAGEN is far superior in terms of speed and accuracy to the standard BWA-GATK implementation, it still suffers from the lack of analysis auditing and tracking to achieve truly standardised and reproducible secondary NGS analyses.
CloudOS brings the power of Illumina’s DRAGEN to your data, in your environment
At Lifebit, we understand the importance of standardisation, reproducibility, auditability and making all stages of analysis FAIR. That is why we always prioritise adding features to the CloudOS platform that will enable users to access the best-in-class tools to standardise all aspects of their analysis. CloudOS makes Illumina’s DRAGEN easily accessible to anyone through the CloudOS Marketplace, at no extra cost, as users only need to cover the standard Illumina DRAGEN-AWS pay-as-you-go fees.
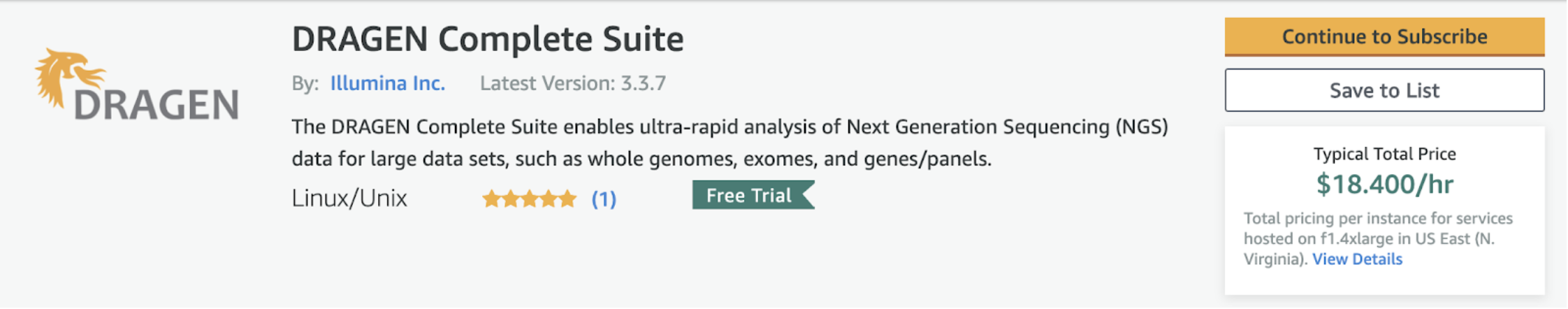
The biggest advantages of running Illumina’s DRAGEN through CloudOS, instead of natively over AWS, are:
- Getting easy access to and deploying Illumina’s DRAGEN over your own AWS cloud
- Managing all data, environment and Illumina’s DRAGEN in one place
- Getting out-of-the-box versioning, 1-click cloning/reproducing and sharing of any analysis performed with Illumina’s DRAGEN
- Enabling real-time monitoring of the analysis progress, owner, resources it uses, its cost, inputs and outputs
- Reducing the cost of running Illumina’s DRAGEN over AWS
When using CloudOS to run Illumina’s DRAGEN, you significantly decrease costs and turnaround times, while at the same time improve the accuracy, trackability and reproducibility of your secondary analyses.
We would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!
Featured news and events
2025-03-26 11:17:46
2025-03-14 15:45:18
2025-03-05 12:49:53
2025-02-27 10:00:00

2025-02-19 13:30:24
2025-02-11 08:39:49
2025-01-30 12:47:38
2025-01-28 08:00:00

2025-01-23 09:07:20

2025-01-08 13:58:41

.png)