

Scaling Reproducible Genomics Analyses for Clinical Environments: The Next Frontier in Healthcare
Lifebit
A bit of background
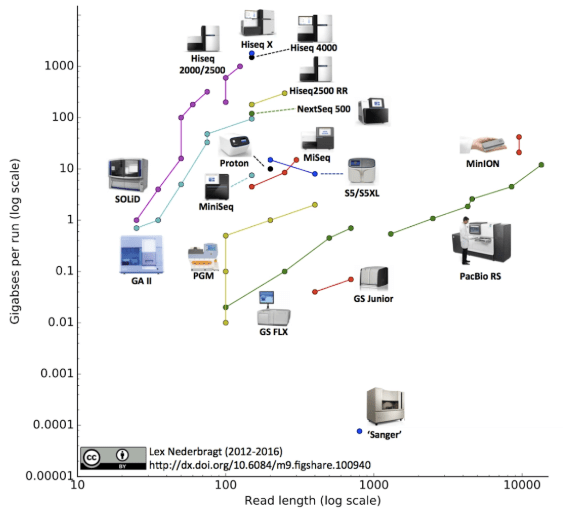
Much of the work in genomics over the last two decades has been focused on the research environment, whether it be sequencing genomes of different species, or identifying changes in the sequence, structure or expression of genomes. The most notable (and most complete) genome sequenced to date is the human genome.
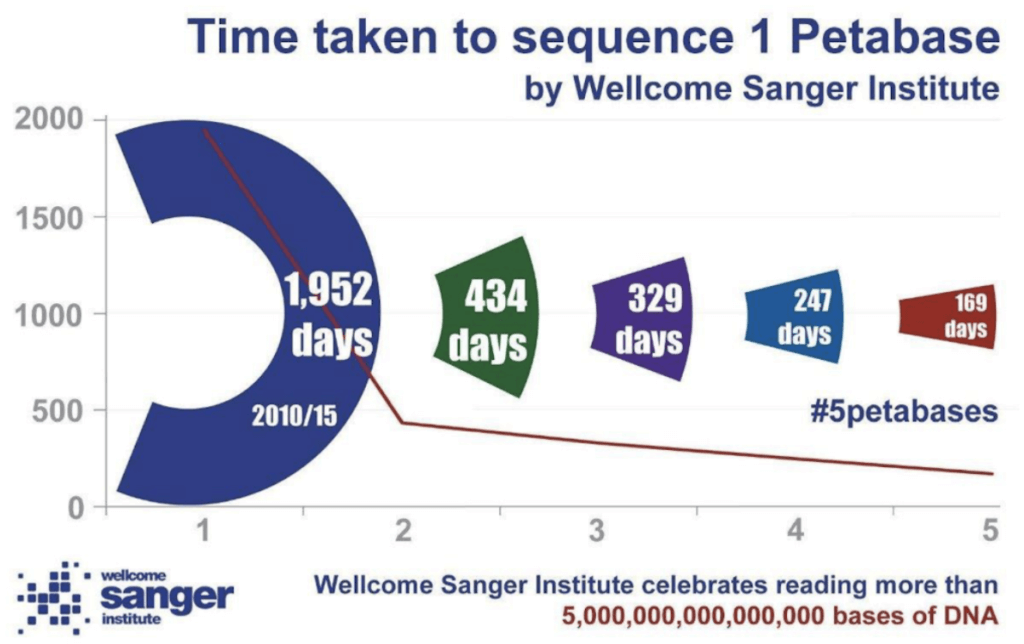
This growth has been driven by technology developments increasing the throughput of sequencing instruments to a point where labs are capable of sequencing hundreds of human genomes in a week. A consequence of this has been the rapid increase in the size of the data being generated. As an example, the Wellcome Sanger Institute in Cambridge recently published an update on their data generation alone:
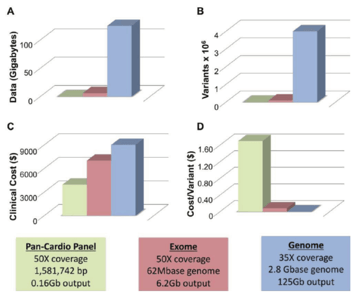
In the background, genetic testing for diagnosing rare disease and cancer has been developing more slowly, partly due to the increased regulatory requirements in a clinical environment1. In addition, the added value of whole genome or exome sequencing against just using targeted gene panels, has not yet been demonstrated, so the extra spending could not be easily justified financially. Even today, many cancer tests will be panels of genes, ranging from a few genes to a few hundred.
Five years ago, the 100,000 Genomes Project was announced by Genomics England with the specific goal of looking at how whole genome sequencing (WGS) could be implemented in a clinical testing environment. It was built upon the work of the Deciphering Developmental Disorders project, which had been using exome sequencing carried out on patients for whom no diagnosis was found by current medical testing.
The current state of clinical genomics
Just recently, NHS announced a truly incredible moment in the history of healthcare: starting on the 1st of October 2018, cancer patients will routinely have their tumours screened for key mutations to facilitate drug treatment choice. What’s more, if that was not enough, the NHS is also planning to have 5,000,000 human genomes sequenced in the next 5 years.
In parallel, industry has begun to move their genomics into the clinical environment. In 2014, Regeneron began a clinical initiative with Geisinger Health to initially sequence 100,000 exomes and share the longitudinal clinical data from these same healthcare members, which currently continues to expand in scale. In 2015, Nelson et al showed that drug targets with genetic evidence were up to 4x more likely to successfully provide marketable drugs3. AstraZeneca announced it wants to sequence 2,000,000 genomes, including the participants in their clinical trials.
In contrast to the research setting, clinical sequencing provides a whole extraa level of regulatory challenges for the collection, storage and analysis of data generated. In addition, there are elements of these analyses that are a key factor in showing the reproducibility of the analysis pipelines and workflows used as part of the analyses. Workflows such as Nextflow have been shown to be highly reproducible4. Use of these workflows and container technologies has allowed easy sharing of pipelines and workflows in research settings. Furthermore, tools like CloudOS allow for a faster implementation and a scalable deployment of these workflows and containers, automating scaling across cloud environments through a web interface and a fully-fledged API.
Looking to the future
As data begins to scale to a point where we are generating exabytes of data per year, it is no longer feasible to rely on internal computing infrastructures. The cloud is coming into its own, and ultimately, we will not be working with just a single cloud environment, but rather, across a multitude of cloud environments and internal servers. To deal with the scale of data, it becomes impractical to move the data to the computing environment, rather than the other way around. So we now need bioinformatic platforms that can bring the computation and analysis tools to the data across multiple cloud environments.
.png?width=624&height=140&name=Screen-Shot-2018-12-21-at-15.39.24%20(1).png)
The rewards of applying these technologies in the clinic can be significant – whether it be for selecting patients for clinical studies, stratifying populations within trials, or retrospectively identifying markers of drug efficacy, driving us closer to personalised treatments5.
References
- Luh, Frank, and Yun Yen. “FDA guidance for next generation sequencing-based testing: balancing regulation and innovation in precision medicine.” NPJ genomic medicine 3.1 (2018): 28.
- Puckelwartz, Megan, and Elizabeth McNally. “Genetic profiling for risk reduction in human cardiovascular disease.” Genes 5.1 (2014): 214-234.
- Nelson, Matthew R., et al. “The support of human genetic evidence for approved drug indications.” Nature genetics 47.8 (2015): 856.
- Di Tommaso, Paolo, et al. “Nextflow enables reproducible computational workflows.” Nature biotechnology 35.4 (2017): 316.
- Mike Furness. “Faster CDx by aligning discovery & clinical data in the regulatory domain.” Pistoia Alliance (2017).
We would like to know what you think! Please fill out the following form or contact us at hello@lifebit.ai. We welcome your comments and suggestions!
Featured news and events
2025-03-26 11:17:46
2025-03-14 15:45:18
2025-03-05 12:49:53
2025-02-27 10:00:00

2025-02-19 13:30:24
2025-01-30 12:47:38

.png)